Hnf1b renal expression directed by a distal enhancer responsive to Pax8
- PMID: 36402859
- PMCID: PMC9675860
- DOI: 10.1038/s41598-022-21171-x
Hnf1b renal expression directed by a distal enhancer responsive to Pax8
Abstract
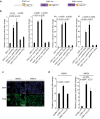
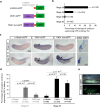
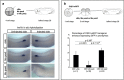
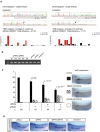
Xenopus provides a simple and efficient model system to study nephrogenesis and explore the mechanisms causing renal developmental defects in human. Hnf1b (hepatocyte nuclear factor 1 homeobox b), a gene whose mutations are the most commonly identified genetic cause of developmental kidney disease, is required for the acquisition of a proximo-intermediate nephron segment in Xenopus as well as in mouse. Genetic networks involved in Hnf1b expression during kidney development remain poorly understood. We decided to explore the transcriptional regulation of Hnf1b in the developing Xenopus pronephros and mammalian renal cells. Using phylogenetic footprinting, we identified an evolutionary conserved sequence (CNS1) located several kilobases (kb) upstream the Hnf1b transcription start and harboring epigenomic marks characteristics of a distal enhancer in embryonic and adult renal cells in mammals. By means of functional expression assays in Xenopus and mammalian renal cell lines we showed that CNS1 displays enhancer activity in renal tissue. Using CRISPR/cas9 editing in Xenopus tropicalis, we demonstrated the in vivo functional relevance of CNS1 in driving hnf1b expression in the pronephros. We further showed the importance of Pax8-CNS1 interaction for CNS1 enhancer activity allowing us to conclude that Hnf1b is a direct target of Pax8. Our work identified for the first time a Hnf1b renal specific enhancer and may open important perspectives into the diagnosis for congenital kidney anomalies in human, as well as modeling HNF1B-related diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Retinoic acid control of pax8 during renal specification of Xenopus pronephros involves hox and meis3.Dev Biol. 2023 Jan;493:17-28. doi: 10.1016/j.ydbio.2022.10.009. Epub 2022 Oct 22. Dev Biol. 2023. PMID: 36279927
-
HNF1B Alters an Evolutionarily Conserved Nephrogenic Program of Target Genes.J Am Soc Nephrol. 2023 Mar 1;34(3):412-432. doi: 10.1681/ASN.2022010076. Epub 2022 Dec 15. J Am Soc Nephrol. 2023. PMID: 36522156 Free PMC article.
-
The nephrogenic potential of the transcription factors osr1, osr2, hnf1b, lhx1 and pax8 assessed in Xenopus animal caps.BMC Dev Biol. 2011 Jan 31;11:5. doi: 10.1186/1471-213X-11-5. BMC Dev Biol. 2011. PMID: 21281489 Free PMC article.
-
HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum.Nat Rev Nephrol. 2015 Feb;11(2):102-12. doi: 10.1038/nrneph.2014.232. Epub 2014 Dec 23. Nat Rev Nephrol. 2015. PMID: 25536396 Review.
-
HNF1B Transcription Factor: Key Regulator in Renal Physiology and Pathogenesis.Int J Mol Sci. 2024 Oct 2;25(19):10609. doi: 10.3390/ijms251910609. Int J Mol Sci. 2024. PMID: 39408938 Free PMC article. Review.
Cited by
-
Sexually dimorphic renal expression of Klotho is directed by a kidney-specific distal enhancer responsive to HNF1b.Res Sq [Preprint]. 2024 Apr 22:rs.3.rs-4188774. doi: 10.21203/rs.3.rs-4188774/v1. Res Sq. 2024. Update in: Commun Biol. 2024 Sep 14;7(1):1142. doi: 10.1038/s42003-024-06855-6. PMID: 38712042 Free PMC article. Updated. Preprint.
-
Sexually dimorphic renal expression of mouse Klotho is directed by a kidney-specific distal enhancer responsive to HNF1b.Commun Biol. 2024 Sep 14;7(1):1142. doi: 10.1038/s42003-024-06855-6. Commun Biol. 2024. PMID: 39277686 Free PMC article.
References
-
- Desgrange A, et al. HNF1B controls epithelial organization and cell polarity during ureteric bud branching and collecting duct morphogenesis. Development. 2017;144:4704–4719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

