Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones
- PMID: 36400772
- PMCID: PMC9674577
- DOI: 10.1038/s41467-022-34875-5
Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones
Abstract
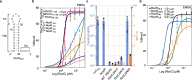
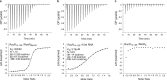
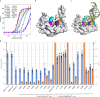
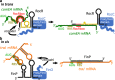
The ProQ/FinO family of RNA binding proteins mediate sRNA-directed gene regulation throughout gram-negative bacteria. Here, we investigate the structural basis for RNA recognition by ProQ/FinO proteins, through the crystal structure of the ProQ/FinO domain of the Legionella pneumophila DNA uptake regulator, RocC, bound to the transcriptional terminator of its primary partner, the sRNA RocR. The structure reveals specific recognition of the 3' nucleotide of the terminator by a conserved pocket involving a β-turn-α-helix motif, while the hairpin portion of the terminator is recognized by a conserved α-helical N-cap motif. Structure-guided mutagenesis reveals key RNA contact residues that are critical for RocC/RocR to repress the uptake of environmental DNA in L. pneumophila. Structural analysis and RNA binding studies reveal that other ProQ/FinO domains also recognize related transcriptional terminators with different specificities for the length of the 3' ssRNA tail.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The Structure of the Human Respiratory Syncytial Virus M2-1 Protein Bound to the Interaction Domain of the Phosphoprotein P Defines the Orientation of the Complex.mBio. 2018 Nov 13;9(6):e01554-18. doi: 10.1128/mBio.01554-18. mBio. 2018. PMID: 30425144 Free PMC article.
-
Legionella pneumophila IrsA, a novel, iron-regulated exoprotein that facilitates growth in low-iron conditions and modulates biofilm formation.Microbiol Spectr. 2025 Jan 7;13(1):e0231324. doi: 10.1128/spectrum.02313-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39612475 Free PMC article.
-
Structure-guided mutagenesis of Henipavirus receptor-binding proteins reveals molecular determinants of receptor usage and antibody-binding epitopes.J Virol. 2024 Mar 19;98(3):e0183823. doi: 10.1128/jvi.01838-23. Epub 2024 Mar 1. J Virol. 2024. PMID: 38426726 Free PMC article.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
Cited by
-
Biochemical and genetic dissection of the RNA-binding surface of the FinO domain of Escherichia coli ProQ.bioRxiv [Preprint]. 2023 Apr 25:2023.04.25.538249. doi: 10.1101/2023.04.25.538249. bioRxiv. 2023. Update in: RNA. 2023 Nov;29(11):1772-1791. doi: 10.1261/rna.079697.123 PMID: 37163069 Free PMC article. Updated. Preprint.
-
Biochemical and genetic dissection of the RNA-binding surface of the FinO domain of Escherichia coli ProQ.RNA. 2023 Nov;29(11):1772-1791. doi: 10.1261/rna.079697.123. Epub 2023 Aug 22. RNA. 2023. PMID: 37607742 Free PMC article.
-
Generation of single-cysteine E. coli ProQ variants to study RNA-protein interaction mechanisms.MicroPubl Biol. 2024 Apr 9;2024:10.17912/micropub.biology.001188. doi: 10.17912/micropub.biology.001188. eCollection 2024. MicroPubl Biol. 2024. PMID: 38660567 Free PMC article.
-
FinO/ProQ-family proteins: an evolutionary perspective.Biosci Rep. 2023 Mar 31;43(3):BSR20220313. doi: 10.1042/BSR20220313. Biosci Rep. 2023. PMID: 36787218 Free PMC article. Review.
-
Antibacterial Ingredients and Modes of the Methanol-Phase Extract from the Fruit of Amomum villosum Lour.Plants (Basel). 2024 Mar 14;13(6):834. doi: 10.3390/plants13060834. Plants (Basel). 2024. PMID: 38592864 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

