Retrotransposon activation during Drosophila metamorphosis conditions adult antiviral responses
- PMID: 36396707
- PMCID: PMC9795486
- DOI: 10.1038/s41588-022-01214-9
Retrotransposon activation during Drosophila metamorphosis conditions adult antiviral responses
Abstract
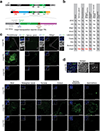
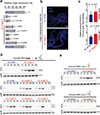
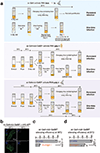
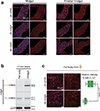
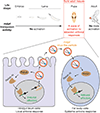
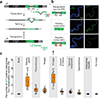
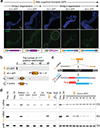
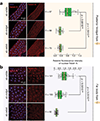
Retrotransposons are one type of mobile genetic element that abundantly reside in the genomes of nearly all animals. Their uncontrolled activation is linked to sterility, cancer and other pathologies, thereby being largely considered detrimental. Here we report that, within a specific time window of development, retrotransposon activation can license the host's immune system for future antiviral responses. We found that the mdg4 (also known as Gypsy) retrotransposon selectively becomes active during metamorphosis at the Drosophila pupal stage. At this stage, mdg4 activation educates the host's innate immune system by inducing the systemic antiviral function of the nuclear factor-κB protein Relish in a dSTING-dependent manner. Consequently, adult flies with mdg4, Relish or dSTING silenced at the pupal stage are unable to clear exogenous viruses and succumb to viral infection. Altogether, our data reveal that hosts can establish a protective antiviral response that endows a long-term benefit in pathogen warfare due to the developmental activation of mobile genetic elements.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures










Comment in
-
Endogenous retrovirus expression during fruitfly metamorphosis enhances adult viral immunity.Nat Genet. 2022 Dec;54(12):1765-1767. doi: 10.1038/s41588-022-01209-6. Nat Genet. 2022. PMID: 36396708 No abstract available.
Similar articles
-
[Precise excision of long terminal repeats of the gypsy (mdg4) retrotransposon of Drosophila melanogaster detected in Escherichia coli cells is explained by its integrase function].Genetika. 2006 Dec;42(12):1656-63. Genetika. 2006. PMID: 17326385 Russian.
-
The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo.EMBO J. 1997 Apr 1;16(7):1732-41. doi: 10.1093/emboj/16.7.1732. EMBO J. 1997. PMID: 9130717 Free PMC article.
-
[Features of the structure of the 7K-copy of Drosophila MDG4 (gypsy) retrotransposon provides evidence that the 7K-subfamily of MDG4 is potentially capable of transposition].Genetika. 1995 Jun;31(6):753-8. Genetika. 1995. PMID: 7635314 Russian.
-
Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila.Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9378-83. doi: 10.1073/pnas.93.18.9378. Proc Natl Acad Sci U S A. 1996. PMID: 8790337 Free PMC article. Review.
-
[Retrotransposon MDG4 and its role in genetic instability of a mutator strain of Drosophila melanogaster].Genetika. 2003 Feb;39(2):164-72. Genetika. 2003. PMID: 12669411 Review. Russian.
Cited by
-
Regulatory logic and transposable element dynamics in nematode worm genomes.bioRxiv [Preprint]. 2024 Sep 16:2024.09.15.613132. doi: 10.1101/2024.09.15.613132. bioRxiv. 2024. PMID: 39345564 Free PMC article. Preprint.
-
Condensin-mediated restriction of retrotransposable elements facilitates brain development in Drosophila melanogaster.Nat Commun. 2024 Mar 28;15(1):2716. doi: 10.1038/s41467-024-47042-9. Nat Commun. 2024. PMID: 38548759 Free PMC article.
-
Dysregulation of innate immune signaling in animal models of Spinal Muscular Atrophy.bioRxiv [Preprint]. 2023 Dec 15:2023.12.14.571739. doi: 10.1101/2023.12.14.571739. bioRxiv. 2023. Update in: BMC Biol. 2024 Apr 25;22(1):94. doi: 10.1186/s12915-024-01888-z PMID: 38168196 Free PMC article. Updated. Preprint.
-
Dysregulation of innate immune signaling in animal models of spinal muscular atrophy.BMC Biol. 2024 Apr 25;22(1):94. doi: 10.1186/s12915-024-01888-z. BMC Biol. 2024. PMID: 38664795 Free PMC article.
-
Insect hormone PTTH regulates lifespan through temporal and spatial activation of NF-κB signaling during metamorphosis.bioRxiv [Preprint]. 2024 Aug 18:2023.09.30.560323. doi: 10.1101/2023.09.30.560323. bioRxiv. 2024. PMID: 37873203 Free PMC article. Preprint.
References
-
- Feschotte C & Gilbert C Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13, 283–96 (2012). - PubMed
-
- Garfinkel DJ, Boeke JD & Fink GR Ty element transposition: reverse transcriptase and virus-like particles. Cell 42, 507–17 (1985). - PubMed
-
- Boeke JD, Garfinkel DJ, Styles CA & Fink GR Ty elements transpose through an RNA intermediate. Cell 40, 491–500 (1985). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

