XAF1 prevents hyperproduction of type I interferon upon viral infection by targeting IRF7
- PMID: 36394357
- PMCID: PMC9827551
- DOI: 10.15252/embr.202255387
XAF1 prevents hyperproduction of type I interferon upon viral infection by targeting IRF7
Abstract
Interferon regulatory factor (IRF) 3 and IRF7 are master regulators of type I interferon (IFN-I)-dependent antiviral innate immunity. Upon viral infection, a positive feedback loop is formed, wherein IRF7 promotes further induction of IFN-I in the later stage. Thus, it is critical to maintain a suitably low level of IRF7 to avoid the hyperproduction of IFN-I. In this study, we find that early expression of IFN-I-dependent STAT1 promotes the expression of XAF1 and that XAF1 is associated specifically with IRF7 and inhibits the activity of XIAP. XAF1-knockout and XIAP-transgenic mice display resistance to viral infection, and this resistance is accompanied by increases in IFN-I production and IRF7 stability. Mechanistically, we find that the XAF1-XIAP axis controls the activity of KLHL22, an adaptor of the BTB-CUL3-RBX1 E3 ligase complex through a ubiquitin-dependent pathway. CUL3-KLHL22 directly targets IRF7 and catalyzes its K48-linked ubiquitination and proteasomal degradation. These findings reveal unexpected functions of the XAF1-XIAP axis and KLHL22 in the regulation of IRF7 stability and highlight an important target for antiviral innate immunity.
Keywords: IFN-I; IRF7; KLHL22; XAF1-XIAP axis; ubiquitination.
© 2022 The Authors.
Figures

- A, B
mRNA levels of IFN‐I (A) and IRF7 (B) in virus‐stimulated WT BMDMs were measured by qRT–PCR at the indicated time points.
- C
WT BMDMs were stimulated with SeV or VSV‐WT (MOI = 1) for 9 h and treated with MG132 2 h before harvest. The indicated proteins in whole‐cell lysates were detected by IB.
- D
WT BMDMs were stimulated with SeV for the indicated time course, and whole‐cell lysates were subjected to IP using an anti‐IRF7 antibody. Cell lysates were also subjected to direct IB, and actin was used as a loading control.
- E
Schematic of the experimental procedure for RNA‐seq and MS.
- F
Left panel, KEGG analysis of IRF7‐interacting proteins as determined by MS; middle panel, Venn diagrams illustrating the overlap between DEGs identified through analysis of the RNA‐seq and MS results; right panel, heatmap showing overlapping DEGs.
- G
Abundant XAF1 mRNA expression was induced by different influenza viruses at an MOI of 1 for 6 h in PBMCs obtained from healthy donors (n = 64) as measured by qRT–PCR.
- H, I
The mRNA (H) and protein (I) levels of XAF1 in virus‐induced WT BMDMs were measured throughout the indicated time course by qRT–PCR and IB, respectively.
- J, K
qRT–PCR analysis of XAF1 mRNA in BMDMs derived from IFNAR‐KO mice (J) and STAT1‐KO mice (K).

HEK293T cells were transfected with IFN‐β luciferase reporters and XAF1‐expressing plasmids and stimulated with SeV or VSV‐WT (MOI = 1). The readouts were normalized to Renilla luciferase activity and are presented as the fold change relative to activity in untransfected cells.
qRT–PCR analysis of IFN‐I induction in WT and XAF1‐deficient BMDMs stimulated with different viruses.
WT or XAF1‐deficient BMDMs were infected with GFP‐expressing VSV (VSV‐GFP) at an MOI of 0.1 for 8 h. The data are presented as representative images and FACS data, showing the infected (GFP+) and total (bright field) cells. Scale bar, 1,000 μm.
qRT–PCR analysis of the mRNA levels of IFN‐I in WT and XAF1‐deficient BMDMs stimulated with distinct TLR agonists.
Some of the LPS‐ or polyI:C‐induced ISGs in WT and XAF1‐deficient BMDMs were measured by qPCR.
mRNA levels of the indicated cytokines in WT and XAF1‐deficient BMDMs activated by distinct TLR agonists.
IFN‐I expression in WT and XAF1‐KO MEFs stimulated with SeV or HSV, evaluated by qPCR.

- A, B
Survival rate (A) and body weight loss (B) were monitored for 8 days (n = 10).
- C
Viral titers in the lung and trachea were quantified by TCID50 assay on day 2 (n = 5).
- D
ELISA of IFN‐I levels in the sera of WT and XAF1MKO mice infected with H1N1 strain PR8 on day 2 (n = 3).
- E
H&E‐stained lung tissue sections 2 days after infection. Inflammation scores are presented in a bar graph (n = 4). Scale bar, 200 μm.
- F–H
Survival rate (F, n = 12) and viral titer (G, n = 4) in the spleen and IFN‐β in the serum (H, n = 4) of WT and XAF1MKO mice intravenously injected with HSV‐1 (3 × 106 PFU per mouse).
- I–K
WT and XAF1MKO mice crossed with IFNAR−/− mice were i.n. infected with a sublethal dose (0.1 HA) of PR8. The survival rate (I, n = 10), viral titer in the lung (J, n = 4), and serum IFN‐β level (K, n = 4) were monitored for the indicated times.

- A
Genotyping PCR of WT (XAF1fl/flLyz2+/+) and myeloid‐conditional XAF1‐KO (XAF1MKO, XAF1fl/flLyz2+/Cre) mice.
- B
qRT–PCR analysis showing clear ablation of XAF1 in the BMDMs of XAF1MKO mice.
- C, D
Flow cytometry analysis of macrophages (CD11b+F4/80+) and neutrophils (CD11b+Ly6G+) in bone marrow (BM) from 6‐ to 8‐week‐old WT and XAF1MKO mice (n = 3).
- E, F
Flow cytometry analysis of the frequencies of distinct immune cells in the spleen (left), naïve and memory T cells (middle), and Treg cells (right) from WT or XAF1MKO mice (n = 3).
- G
Volcano plot illustrating the upregulated and downregulated DEGs in XAF1‐deficient BMDMs compared with WT littermates after 6 h of LPS stimulation. The KEGG analysis showed that the upregulated DEGs were enriched in biological processes.
- H
qRT–PCR analysis of ARG1, MRC1, and YM1 in IL‐4‐treated M2 macrophages in WT and XAF1‐deficient BMDMs.
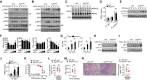
- A, B
Virus‐induced phosphorylation of TBK1 and IRF3 in cytoplasmic (CE) and nuclear (NE) extracts of WT and XAF1‐deficient BMDMs was measured by IB.
- C
Monomeric and dimeric IRF3 expression (top blot) in whole‐cell lysates of WT and XAF1‐deficient BMDMs stimulated with VSV‐WT (MOI = 1) was measured by IB.
- D
ChIP assays were performed, and the results were quantified by qRT–PCR to detect IRF3 binding to an ISRE in WT and XAF1‐deficient BMDMs activated by VSV‐WT (MOI = 1) for 9 h.
- E
IB was performed to determine the VSV‐induced phosphorylation level of STAT1.
- F
HEK293T cells were transfected with IFN‐β luciferase reporters in the presence (+) or absence (−) of the indicated XAF1 expression plasmids. Luciferase assays were used to determine the fold change in expression with respect to the empty vector group 36 h after transfection.
- G
qRT–PCR analysis of Irf7 mRNA in WT and XAF1‐deficient BMDMs stimulated with distinct virus infections.
- H, I
IB analysis of VSV‐ or HSV‐1‐induced IRF7 expression as measured in whole‐cell lysates of WT and XAF1‐deficient BMDMs.
- J
ChIP assays were performed and quantified by qRT–PCR to detect IRF7 binding to an ISRE in WT and XAF1‐deficient BMDMs stimulated with VSV‐WT (MOI = 1) or HSV‐1.
- K–N
WT and XAF1MKO mice crossed with mice with an IRF7−/− background were i.n. infected with a sublethal dose (0.1 hyaluronic acid [HA]) of the H1N1 strain PR8. The survival rate (K, n = 15), viral titer in the lung (L, n = 5), serum IFN‐β level (M, n = 5), and intensity of H&E‐stained lung tissue sections (N, n = 5) were determined at the indicated time points. Scale bar, 200 μm.

- A, B
Body weight loss (A) and stool diarrhea and hematochezia scores (B) in WT and XAF1MKO mice of the DSS‐induced colitis model.
- C, D
Colon length (C) and proinflammatory cytokine production (D) of DSS‐treated WT and XAF1MKO mice on day 8.
- E, F
The indicated proteins in cytoplasmic (CE) and nuclear extracts (NE) of WT and XAF1‐deficient BMDMs were detected by IB.
- G
The activation of MAPKs in total cell lysates was measured by IB.

- A
After treatment with CQ or MG132, IRF7 in nuclear extracts of WT and XAF1‐deficient BMDMs was detected by IB.
- B, C
WT and XAF1‐deficient BMDMs were infected with VSV‐WT (MOI = 1) and treated with MG132 for 2 h before harvest. Whole‐cell lysates (WLs) were subjected to IP using an anti‐IRF7 antibody, and the indicated proteins were detected by IB.
- D
HEK293T cells were cotransfected with XAF1 and distinct IRF‐expressing plasmids. IB HA was performed followed by IP with an anti‐FLAG antibody.
- E
The interaction between XAF1 and IRF7 was assessed in WT BMDMs stimulated by VSV‐WT (MOI = 1). WLs were subjected to IP using an anti‐IRF7 or anti‐IgG antibody and then to IB and detected with anti‐XAF1 antibody.
- F
The associations between XAF1 and various IRF7 truncation mutants were detected through IP and IB of the transfected HEK293T cells.
- G, H
The associations between IRF7 and various XAF1 truncation mutants were detected through IP and IB.
- I
qRT–PCR analysis of the indicated genes in XAF1‐deficient MEFs expressing XAF1WT and XAF1ΔZF6 and subjected to the indicated virus stimulation.

- A
Model diagram showing the potential regulatory mechanisms involving XAF1, XIAP, and IRF7.
- B
qRT–PCR analysis of XIAP expression in WT and XAF1‐deficient BMDMs stimulated with IFN‐β.
- C
HEK293T cells were transfected with IFN‐β luciferase reporters in the presence (+) or absence (−) of the indicated XIAP expression plasmids. Luciferase assays were performed, and the results are reported as fold changes with respect to the empty vector group 36 h after transfection.
- D, E
qRT–PCR analysis of IFN‐I expression in WT or XIAPTg BMDMs stimulated with distinct viruses (D) or TLR agonists (E).
- F, G
The virus‐induced expression of IRF7 in WT and XIAPTg BMDMs was measured by IB.
- H
WT and XIAPTg BMDMs were infected with VSV‐WT (MOI = 1) and treated with MG132 2 h before harvest. Whole‐cell lysates were subjected to IP using an anti‐IRF7 antibody.
- I
HEK293T cells were transfected with FLAG‐tagged ubiquitin and the indicated expression plasmids. The ubiquitination level of IRF7 was determined by IB, and total cell lysates were also subjected to direct IB.
- J
HEK293T cells were cotransfected with XAF1 and distinct IRF‐expressing plasmids. IBHA was performed, followed by IP with an anti‐FLAG antibody.
- K
The interaction between XIAP and IRF7 was assessed in XIAP‐overexpressing MEFs activated by SeV. Whole‐cell lysates (WLs) were subjected to IP using an anti‐FLAG‐XIAP or anti‐IgG antibody and then subjected to IB and detected with the anti‐IRF7 antibody.
- L
The associations between IRF7 and various XIAP truncation mutants were detected through IP and IB.

- A–F
Flow cytometry analysis of the absolute numbers of different immune cells in the spleen (A, B, upper), inguinal lymph nodes (C, D, middle), and thymus (E, F, lower) from WT and XAF1MKO mice (n = 3).
- G, H
Flow cytometry analysis of macrophages (CD11b+F4/80+) and neutrophils (CD11b+Ly6G+) in bone marrow (BM) from 6‐ to 8‐week‐old WT and XAF1MKO mice (n = 3).

- A
The list of ubiquitination (Ub)‐related proteins that interact with IRF7 according to the MS results.
- B
HEK293T cells were transfected with IRF7 and the indicated ubiquitin‐related expression plasmid. IB was performed to detect FLAG, followed by IP with an anti‐hyaluronic acid (HA) antibody.
- C, D
HEK293T cells were transfected with IFN‐β luciferase reporters and the indicated expression plasmids. Luciferase assays were performed to determine the fold changes with respect to the empty vector group.
- E
qRT–PCR analysis of IFN‐I in WT and KLHL22‐KO MEFs stimulated with polyI:C or SeV. The data are presented as the fold change relative to the Actb mRNA level.
- F
Immunoblot analysis of SeV‐induced IRF7 in whole‐cell lysates (WLs) of WT or KLHL22‐KO MEFs.
- G
After treatment with MG132, IRF7 was isolated by IP from WLs of KLHL22‐KO MEFs and subjected to IB using anti‐K48‐ubiquitin. Total cell lysates were also subjected to direct IB.
- H
HEK293T cells were transfected with IRF7 and FLAG‐K48R‐ubiquitin or FLAG‐ubiquitin in the presence (+) or absence (−) of KLHL22 expression plasmids. HA‐tagged IRF7 was isolated by IP, and the ubiquitination level was then measured by IB.
- I
HEK293T cells were transfected with IRF7 and distinct KLHL22 truncation mutants. IB of FLAG was performed, followed by IP with an anti‐HA antibody.
- J
HEK293T cells were transfected with KLHL22, FLAG‐K48‐ubiquitin, and various IRF7 point mutants. After treatment with MG132, IB of K48‐Ub was performed, followed by IP with an anti‐HA antibody.
- K
HEK293T cells were transfected with IFN‐α4 luciferase reporters and the indicated expression plasmids. Luciferase assays were performed to determine fold changes with respect to the empty vector group.

- A
HEK293T cells were transfected with IRF7 and the indicated expression plasmids. After treatment with MG132, the indicated proteins were detected by IB.
- B, C
IB and qRT–PCR analysis showing specific ablation of KLHL22 in KLHL22‐KO MEFs generated by CRISPR.
- D
After treatment with MG132, IRF7 was isolated by IP from whole‐cell lysates (WLs) of KLHL22‐KO MEFs and subjected to IB using anti‐ubiquitin. Total cell lysates were also subjected to direct IB.
- E
HEK293T cells were transfected with IRF7 and FLAG‐K48‐ubiquitin in the presence (+) or absence (−) of KLHL22 expression plasmids. After pretreatment with MG132 or CQ, HA‐tagged IRF7 was isolated by IP, and the ubiquitination level was then measured by IB.
- F
HEK293T cells were transfected with multiple ubiquitin mutants (mutations at K6, K11, K27, K29, K33, K48, and K63) and the indicated expression plasmids. HA‐tagged IRF7 was isolated by IP, and the ubiquitination level was then measured by IB.
- G, H
HEK293T cells were transfected with HA‐tagged IRF7 and the inactive mutant variants (S477 and 479A) whose phosphorylation sites at serine 477 and 479 were mutated from serine to alanine. HA‐tagged IRF7 was isolated by IP, and the ubiquitination level was then measured by IB.
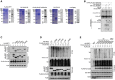
Purification of recombinant proteins was performed by Coomassie blue staining and IB.
CUL3‐KLHL22 E3 ligase catalyzed IRF7 ubiquitination in a cell‐free system. Ubiquitin and recombinant FLAG‐IRF7 were incubated with recombinant CUL3–RBX1 and KLHL22 or KLHL22‐Δ6xKelch.
The associations between KLHL22 and various truncations of IRF7 were detected through the indicated IP and IB analyses.
HEK293T cells were transfected with KLHL22 and IRF7 variants. IB of FLAG was performed followed by IP with an anti‐HA antibody.
HEK293T cells were transfected with KLHL22, FLAG‐ubiquitin, and various IRF7 point mutants. IB of FLAG was performed, followed by IP with an anti‐HA antibody. The data are representative of at least three biologically independent experiments.
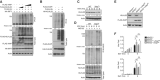
HEK293T cells were transfected with FLAG‐tagged ubiquitin and the indicated expression plasmids. The ubiquitination levels of IRF7 were measured by IB. Cell lysates were subjected to direct IB.
HEK293T cells were transfected with KLHL22 in the presence (+) or absence (−) of XIAP. IB of FLAG was performed followed by IP with an anti‐hyaluronic acid (HA) antibody.
IB of VSV‐induced KLHL22 expression in whole‐cell lysates (WLs) of WT and Xiap Tg mice.
After treatment with MG132, KLHL22 was isolated by IP from WLs of WT and Xiap Tg mice and subjected to IB using an anti‐ubiquitin or anti‐K48‐ubiquitin antibody. Protein lysates were also subjected to direct IB.
IB showed the ablation of KLHL22 and the overexpression of FLAG‐XIAP in KLHL22‐KO MEFs expressing XIAP (KLHL22−/−XIAPOE).
qRT–PCR analysis of IFN‐I in WT and KLHL22‐KO MEFs expressing XIAP and subjected to SeV stimulation.
Similar articles
-
Porcine deltacoronavirus nucleocapsid protein antagonizes JAK-STAT signaling pathway by targeting STAT1 through KPNA2 degradation.J Virol. 2024 Jul 23;98(7):e0033424. doi: 10.1128/jvi.00334-24. Epub 2024 Jun 3. J Virol. 2024. PMID: 38829137 Free PMC article.
-
The antiviral activity of myricetin against pseudorabies virus through regulation of the type I interferon signaling pathway.J Virol. 2024 Nov 27:e0156724. doi: 10.1128/jvi.01567-24. Online ahead of print. J Virol. 2024. PMID: 39601590
-
The Role of REC8 in the Innate Immune Response to Viral Infection.J Virol. 2022 Mar 23;96(6):e0217521. doi: 10.1128/jvi.02175-21. Epub 2022 Feb 2. J Virol. 2022. PMID: 35107381 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Aug 27;8:CD007807. doi: 10.1002/14651858.CD007807.pub4 PMID: 28752910 Free PMC article. Updated. Review.
Cited by
-
XAF1 promotes osteoclast apoptosis by antagonizing the XIAP-caspase axis.J Orthop Translat. 2024 Jun 7;47:15-28. doi: 10.1016/j.jot.2024.05.001. eCollection 2024 Jul. J Orthop Translat. 2024. PMID: 38957269 Free PMC article.
-
IRF7: role and regulation in immunity and autoimmunity.Front Immunol. 2023 Aug 10;14:1236923. doi: 10.3389/fimmu.2023.1236923. eCollection 2023. Front Immunol. 2023. PMID: 37638030 Free PMC article. Review.
-
De novo variants in immune regulatory genes in Down syndrome regression disorder.J Neurol. 2024 Aug;271(8):5567-5576. doi: 10.1007/s00415-024-12521-y. Epub 2024 Jun 22. J Neurol. 2024. PMID: 38909119 Free PMC article.
-
Comparative Study on the Mechanism of Macrophage Activation Induced by Polysaccharides from Fresh and Dried Longan.Nutrients. 2024 May 28;16(11):1654. doi: 10.3390/nu16111654. Nutrients. 2024. PMID: 38892587 Free PMC article.
-
XAF1 promotes colorectal cancer metastasis via VCP-RNF114-JUP axis.J Cell Biol. 2024 Feb 5;223(2):e202303015. doi: 10.1083/jcb.202303015. Epub 2023 Dec 14. J Cell Biol. 2024. PMID: 38095639 Free PMC article.
References
-
- Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H (1996) Pip, a lymphoid‐restricted IRF, contains regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev 10: 2335–2347 - PubMed
-
- Chen J, Ou Y, Yang Y, Li W, Xu Y, Xie Y, Liu Y (2018) KLHL22 activates amino‐acid‐dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 557: 585–589 - PubMed
-
- Dusheiko G (1997) Side effects of alpha interferon in chronic hepatitis C. Hepatology 26: 112S–121S - PubMed
-
- Higgs R, Jefferies CA (2008) Targeting IRFs by ubiquitination: regulating antiviral responses. Biochem Soc Trans 36: 453–458 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

