MatchTope: A tool to predict the cross reactivity of peptides complexed with Major Histocompatibility Complex I
- PMID: 36389840
- PMCID: PMC9650389
- DOI: 10.3389/fimmu.2022.930590
MatchTope: A tool to predict the cross reactivity of peptides complexed with Major Histocompatibility Complex I
Abstract
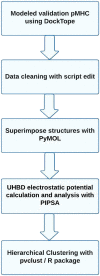
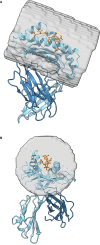
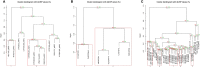
The therapeutic targeting of the immune system, for example in vaccinology and cancer treatment, is a challenging task and the subject of active research. Several in silico tools used for predicting immunogenicity are based on the analysis of peptide sequences binding to the Major Histocompatibility Complex (pMHC). However, few of these bioinformatics tools take into account the pMHC three-dimensional structure. Here, we describe a new bioinformatics tool, MatchTope, developed for predicting peptide similarity, which can trigger cross-reactivity events, by computing and analyzing the electrostatic potentials of pMHC complexes. We validated MatchTope by using previously published data from in vitro assays. We thereby demonstrate the strength of MatchTope for similarity prediction between targets derived from several pathogens as well as for indicating possible cross responses between self and tumor peptides. Our results suggest that MatchTope can enhance and speed up future studies in the fields of vaccinology and cancer immunotherapy.
Keywords: T-cell; T-cell response; cross reactivity; cross reactivity prediction; immunoinformatic.
Copyright © 2022 Mendes, de Souza Bragatte, Vianna, de Freitas, Pöhner, Richter, Wade, Salzano and Vieira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Interpreting T-Cell Cross-reactivity through Structure: Implications for TCR-Based Cancer Immunotherapy.Front Immunol. 2017 Oct 4;8:1210. doi: 10.3389/fimmu.2017.01210. eCollection 2017. Front Immunol. 2017. PMID: 29046675 Free PMC article.
-
T cell receptor/peptide/MHC molecular characterization and standardized pMHC contact sites in IMGT/3Dstructure-DB.In Silico Biol. 2005;5(5-6):505-28. In Silico Biol. 2005. PMID: 16268793
-
Structural Prediction of Peptide-MHC Binding Modes.Methods Mol Biol. 2022;2405:245-282. doi: 10.1007/978-1-0716-1855-4_13. Methods Mol Biol. 2022. PMID: 35298818 Review.
-
MPID-T: database for sequence-structure-function information on T-cell receptor/peptide/MHC interactions.Appl Bioinformatics. 2006;5(2):111-4. doi: 10.2165/00822942-200605020-00005. Appl Bioinformatics. 2006. PMID: 16722775
-
The use of peptide-major-histocompatibility-complex multimers in type 1 diabetes mellitus.J Diabetes Sci Technol. 2012 May 1;6(3):515-24. doi: 10.1177/193229681200600305. J Diabetes Sci Technol. 2012. PMID: 22768881 Free PMC article. Review.
Cited by
-
CrossDome: an interactive R package to predict cross-reactivity risk using immunopeptidomics databases.Front Immunol. 2023 Jun 12;14:1142573. doi: 10.3389/fimmu.2023.1142573. eCollection 2023. Front Immunol. 2023. PMID: 37377956 Free PMC article.
-
HLA3DB: comprehensive annotation of peptide/HLA complexes enables blind structure prediction of T cell epitopes.Nat Commun. 2023 Oct 10;14(1):6349. doi: 10.1038/s41467-023-42163-z. Nat Commun. 2023. PMID: 37816745 Free PMC article.
-
MORITS: An improved method to predict peptides from heterologous proteins that are recognized by the same T-cell receptor.Sci Rep. 2024 Apr 8;14(1):8255. doi: 10.1038/s41598-024-58350-x. Sci Rep. 2024. PMID: 38589549 Free PMC article.
-
FASTMAP-a flexible and scalable immunopeptidomics pipeline for HLA- and antigen-specific T-cell epitope mapping based on artificial antigen-presenting cells.Front Immunol. 2024 May 8;15:1386160. doi: 10.3389/fimmu.2024.1386160. eCollection 2024. Front Immunol. 2024. PMID: 38779658 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

