Structural consequences of sequence variation in mammalian prion β2α2 loop segments
- PMID: 36389229
- PMCID: PMC9645039
- DOI: 10.3389/fnins.2022.960322
Structural consequences of sequence variation in mammalian prion β2α2 loop segments
Abstract
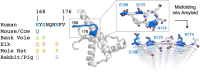
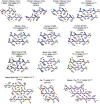
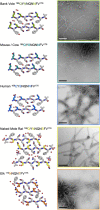
Sequence variation in the β2α2 loop, residues 165-175 of the mammalian prion protein (PrP), influences its structure. To better understand the consequences of sequence variation in this region of the protein, we biochemically and biophysically interrogate natural and artificial sequence variants of the β2α2 loop of mammalian PrP. Using microcrystal electron diffraction (MicroED), we determine atomic resolution structures of segments encompassing residues 168-176 from the β2α2 loop of PrP with sequences corresponding to human, mouse/cow, bank vole/hamster, rabbit/pig/guinea pig, and naked mole rat (elk-T174S) β2α2 loops, as well as synthetic β2α2 loop sequences. This collection of structures presents two dominant amyloid packing polymorphisms. In the first polymorph, denoted "clasped", side chains within a sheet form polar clasps by facing each other on the same strand, exemplified by the mouse/cow, human, and bank vole/hamster sequences. Because its stability is derived from within a strand and through polar ladders within a sheet, the sequence requirements for the mating strand are less restrictive. A second polymorph, denoted "interdigitated," has sidechains interdigitate across mating sheets, exemplified by the elk, naked mole rat (elk T174S), and rabbit sequences. The two types of packing present distinct networks of stabilizing hydrogen bonds. The identity of residue 174 appears to strongly influence the packing adopted in these peptides, but consideration of the overall sequence of a given segment is needed to understand the stability of its assemblies. Incorporation of these β2α2 loop sequences into an 85 residue recombinant segment encoding wild-type bank vole PrP94-178 demonstrates that even single residue substitutions could impact fibril morphology as evaluated by negative stain electron microscopy. This is in line with recent findings supporting the accessibility of different structural geometries by varied mammalian prion sequences, and indicates that sequence-specific polymorphisms may be influenced by residues in the β2α2 loop.
Keywords: cryoEM; microED; prion; structure; transmission.
Copyright © 2022 Glynn, Hernandez, Gallagher-Jones, Miao, Sigurdson and Rodriguez.
Conflict of interest statement
JR is an equity stakeholder in Medstruc Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Asparagine and glutamine ladders promote cross-species prion conversion.J Biol Chem. 2017 Nov 17;292(46):19076-19086. doi: 10.1074/jbc.M117.794107. Epub 2017 Sep 20. J Biol Chem. 2017. PMID: 28931606 Free PMC article.
-
The polar clasps of a bank vole PrP(168-176) prion protofibril revisiting.J Mol Model. 2019 Apr 2;25(5):108. doi: 10.1007/s00894-019-3981-z. J Mol Model. 2019. PMID: 30937536
-
NMR structure of the bank vole prion protein at 20 degrees C contains a structured loop of residues 165-171.J Mol Biol. 2008 Nov 7;383(2):306-12. doi: 10.1016/j.jmb.2008.08.045. Epub 2008 Aug 26. J Mol Biol. 2008. PMID: 18773909
-
Molecular structures of amyloid and prion fibrils: consensus versus controversy.Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review.
-
Structural factors underlying the species barrier and susceptibility to infection in prion disease.Biochem Cell Biol. 2010 Apr;88(2):195-202. doi: 10.1139/o09-172. Biochem Cell Biol. 2010. PMID: 20453922 Review.
References
-
- Apostol M. I., Sawaya M. R., Cascio D., Eisenberg D. (2010). Crystallographic studies of prion protein (prp) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease. J. Biol. Chem. 285 29671–29675. 10.1074/jbc.C110.158303 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

