Rhizospheric microbial consortium of Lilium lancifolium Thunb. causes lily root rot under continuous cropping system
- PMID: 36386686
- PMCID: PMC9645529
- DOI: 10.3389/fmicb.2022.981615
Rhizospheric microbial consortium of Lilium lancifolium Thunb. causes lily root rot under continuous cropping system
Abstract
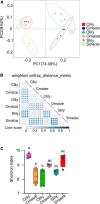
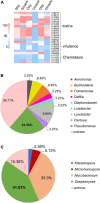
Tiger lily (Lilium lancifolium Thunb.) is a cash crop with a long history of cultivation in China. Its roots have long been used as a valuable component of Chinese medicine. Continuous cropping, the conventional planting approach for tiger lily, often leads to severe root rot disease, but it is not yet clear how this planting method leads to root rot. In this study, we analyzed the rhizosphere microbiome and predicted microbial protein function in tiger lily planted with the continuous cropping method in three different geological types of soil. In order to explore the specific rhizosphere microbiota triggering root rot disease, tiger lily was compared to maize grown in a similar system, which showed no disease development. An analysis of the chemical elements in the soil revealed that the Pseudomonas and Streptomyces genera, with pathogenic functions, were dominant in the tiger lily rhizosphere. The lower soil pH of tiger lily compared to maize supports the accumulation of pathogenic bacteria in the tiger lily rhizosphere. Meanwhile, we discovered that bacteria of the Flavobacterium genus, with their predicted phosphate transport function, specifically accumulated in the maize rhizosphere. Our findings suggest that Pseudomonas and Streptomyces bacteria may result in continuous cropping-induced root rot disease in tiger lily and that Flavobacterium could serve to protect maize from pathogenic bacteria.
Keywords: continuous cropping; geological soil; lily root rot disease; microbiota; rhizosphere.
Copyright © 2022 Dai, Singh, Gong, Tang, Peng, Zhang, Wu, Zhang and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Earthworm activity optimized the rhizosphere bacterial community structure and further alleviated the yield loss in continuous cropping lily (Lilium lancifolium Thunb.).Sci Rep. 2021 Oct 21;11(1):20840. doi: 10.1038/s41598-021-99597-y. Sci Rep. 2021. PMID: 34675325 Free PMC article.
-
Effects of lily/maize intercropping on rhizosphere microbial community and yield of Lilium davidii var. unicolor.J Basic Microbiol. 2018 Oct;58(10):892-901. doi: 10.1002/jobm.201800163. Epub 2018 Aug 12. J Basic Microbiol. 2018. PMID: 30101457
-
Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field.World J Microbiol Biotechnol. 2016 Jun;32(6):95. doi: 10.1007/s11274-016-2051-2. Epub 2016 Apr 27. World J Microbiol Biotechnol. 2016. PMID: 27116961
-
Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous cropping practices.J Basic Microbiol. 2017 Apr;57(4):337-344. doi: 10.1002/jobm.201600464. Epub 2017 Jan 6. J Basic Microbiol. 2017. PMID: 28060404
-
Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years.Front Microbiol. 2023 Sep 7;14:1203796. doi: 10.3389/fmicb.2023.1203796. eCollection 2023. Front Microbiol. 2023. PMID: 37744902 Free PMC article. Review.
Cited by
-
Structure and function of rhizosphere soil microbial communities associated with root rot of Knoxia roxburghii.Front Microbiol. 2024 Jul 18;15:1424633. doi: 10.3389/fmicb.2024.1424633. eCollection 2024. Front Microbiol. 2024. PMID: 39091303 Free PMC article.
-
Integrated metagenomics and metabolomics analysis reveals changes in the microbiome and metabolites in the rhizosphere soil of Fritillaria unibracteata.Front Plant Sci. 2023 Aug 4;14:1223720. doi: 10.3389/fpls.2023.1223720. eCollection 2023. Front Plant Sci. 2023. PMID: 37600181 Free PMC article.
-
A Systematic Review on the Continuous Cropping Obstacles and Control Strategies in Medicinal Plants.Int J Mol Sci. 2023 Aug 5;24(15):12470. doi: 10.3390/ijms241512470. Int J Mol Sci. 2023. PMID: 37569843 Free PMC article. Review.
References
-
- Abadi V. A. J. M., Sepehri M., Rahmani H. A., Zarei M., Ronaghi A., Taghavi S. M., et al. (2020). Role of Dominant phyllosphere bacteria with plant growth–promoting characteristics on growth and nutrition of Maize (Zea mays L.). J. Soil Sci. Plant Nutr. 20 2348–2363.
LinkOut - more resources
Full Text Sources

