Quantitation of SARS-CoV-2 neutralizing antibodies with a virus-free, authentic test
- PMID: 36382127
- PMCID: PMC9645495
- DOI: 10.1093/pnasnexus/pgac045
Quantitation of SARS-CoV-2 neutralizing antibodies with a virus-free, authentic test
Erratum in
-
Correction to: Quantitation of SARS-CoV-2 neutralizing antibodies with a virus-free, authentic test.PNAS Nexus. 2024 Jan 30;3(1):pgad477. doi: 10.1093/pnasnexus/pgad477. eCollection 2024 Jan. PNAS Nexus. 2024. PMID: 38292538 Free PMC article.
Abstract
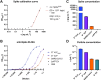
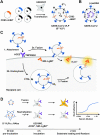
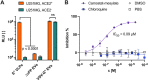
Neutralizing antibodies (NAbs), and their concentration in sera of convalescents and vaccinees are a correlate of protection from COVID-19. The antibody concentrations in clinical samples that neutralize SARS-CoV-2 are difficult and very cumbersome to assess with conventional virus neutralization tests (cVNTs), which require work with the infectious virus and biosafety level 3 containment precautions. Alternative virus neutralization tests currently in use are mostly surrogate tests based on direct or competitive enzyme immunoassays or use viral vectors with the spike protein as the single structural component of SARS-CoV-2. To overcome these obstacles, we developed a virus-free, safe and very fast (4.5 h) in vitro diagnostic test based on engineered yet authentic SARS-CoV-2 virus-like-particles (VLPs). They share all features of the original SARS-CoV-2 but lack the viral RNA genome and thus are non-infectious. NAbs induced by infection or vaccination, but also potentially neutralizing monoclonal antibodies can be reliably quantified and assessed with ease and within hours with our test, because they interfere and block the ACE2-mediated uptake of VLPs by recipient cells. Results from the VLP neutralization test (VLPNT) showed excellent specificity and sensitivity and correlated very well with a cVNT using fully infectious SARS-CoV-2. The results also demonstrated the reduced neutralizing capacity of COVID-19 vaccinee sera against variants of concern of SARS-CoV-2 including omicron B.1.1.529, BA.1.
Keywords: Omicron; SARS-CoV-2; diagnostics; virus neutralization test; virus-like particle.
Conflict of interest statement
Competing interests The authors D.P., R.Z., and W.H. report that the Helmholtz Zentrum Muenchen has filed a patent application relating to SARS-CoV-2 virus neutralization assays. The application lists the authors as inventors.
Figures




Similar articles
-
Comparative analysis of the neutralizing activity against SARS-CoV-2 Wuhan-Hu-1 strain and variants of concern: Performance evaluation of a pseudovirus-based neutralization assay.Front Immunol. 2022 Sep 26;13:981693. doi: 10.3389/fimmu.2022.981693. eCollection 2022. Front Immunol. 2022. PMID: 36225911 Free PMC article.
-
Reduced Binding between Omicron B.1.1.529 and the Human ACE2 Receptor in a Surrogate Virus Neutralization Test for SARS-CoV-2.Viruses. 2023 May 30;15(6):1280. doi: 10.3390/v15061280. Viruses. 2023. PMID: 37376580 Free PMC article.
-
SARS-CoV-2 vaccination of convalescents boosts neutralization capacity against Omicron subvariants BA.1, BA.2 and BA.5 and can be predicted by anti-S antibody concentrations in serological assays.Front Immunol. 2023 Apr 25;14:1170759. doi: 10.3389/fimmu.2023.1170759. eCollection 2023. Front Immunol. 2023. PMID: 37180152 Free PMC article.
-
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone.Int J Mol Sci. 2022 Jul 12;23(14):7675. doi: 10.3390/ijms23147675. Int J Mol Sci. 2022. PMID: 35887023 Free PMC article.
-
Quantifying Absolute Neutralization Titers against SARS-CoV-2 by a Standardized Virus Neutralization Assay Allows for Cross-Cohort Comparisons of COVID-19 Sera.mBio. 2021 Feb 16;12(1):e02492-20. doi: 10.1128/mBio.02492-20. mBio. 2021. PMID: 33593976 Free PMC article.
Cited by
-
Light-Induced Transformation of Virus-Like Particles on TiO2.ACS Appl Mater Interfaces. 2024 Jul 17;16(28):37275-37287. doi: 10.1021/acsami.4c07151. Epub 2024 Jul 3. ACS Appl Mater Interfaces. 2024. PMID: 38959130 Free PMC article.
-
Pseudotyping Improves the Yield of Functional SARS-CoV-2 Virus-like Particles (VLPs) as Tools for Vaccine and Therapeutic Development.Int J Mol Sci. 2023 Sep 27;24(19):14622. doi: 10.3390/ijms241914622. Int J Mol Sci. 2023. PMID: 37834067 Free PMC article.
-
VLP-ELISA for the Detection of IgG Antibodies against Spike, Envelope, and Membrane Antigens of SARS-CoV-2 in Indian Population.Vaccines (Basel). 2023 Mar 27;11(4):743. doi: 10.3390/vaccines11040743. Vaccines (Basel). 2023. PMID: 37112655 Free PMC article.
-
MDM2 Influences ACE2 Stability and SARS-CoV-2 Uptake.Viruses. 2023 Aug 18;15(8):1763. doi: 10.3390/v15081763. Viruses. 2023. PMID: 37632105 Free PMC article.
-
SARS-CoV-2 and Epstein-Barr Virus-like Particles Associate and Fuse with Extracellular Vesicles in Virus Neutralization Tests.Biomedicines. 2023 Oct 25;11(11):2892. doi: 10.3390/biomedicines11112892. Biomedicines. 2023. PMID: 38001893 Free PMC article.

