RNA Metabolism in T Lymphocytes
- PMID: 36381959
- PMCID: PMC9634142
- DOI: 10.4110/in.2022.22.e39
RNA Metabolism in T Lymphocytes
Abstract
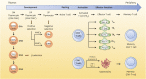
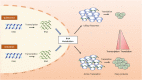
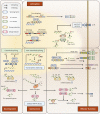
RNA metabolism plays a central role in regulating of T cell-mediated immunity. RNA processing, modifications, and regulations of RNA decay influence the tight and rapid regulation of gene expression during T cell phase transition. Thymic selection, quiescence maintenance, activation, differentiation, and effector functions of T cells are dependent on selective RNA modulations. Recent technical improvements have unveiled the complex crosstalk between RNAs and T cells. Moreover, resting T cells contain large amounts of untranslated mRNAs, implying that the regulation of RNA metabolism might be a key step in controlling gene expression. Considering the immunological significance of T cells for disease treatment, an understanding of RNA metabolism in T cells could provide new directions in harnessing T cells for therapeutic implications.
Keywords: Cellular; Immunity; RNA; RNA metabolism; T-lymphocytes.
Copyright © 2022. The Korean Association of Immunologists.
Conflict of interest statement
Conflict of Interest: The authors declare no potential conflicts of interest.
Figures
Similar articles
-
Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection.Curr Top Microbiol Immunol. 2005;293:43-71. doi: 10.1007/3-540-27702-1_3. Curr Top Microbiol Immunol. 2005. PMID: 15981475 Review.
-
Role of STAT3 in CD4+CD25+FOXP3+ regulatory lymphocyte generation: implications in graft-versus-host disease and antitumor immunity.J Immunol. 2007 Dec 1;179(11):7593-604. doi: 10.4049/jimmunol.179.11.7593. J Immunol. 2007. PMID: 18025205
-
Regulation of Early Lymphocyte Development via mRNA Decay Catalyzed by the CCR4-NOT Complex.Front Immunol. 2021 Jul 19;12:715675. doi: 10.3389/fimmu.2021.715675. eCollection 2021. Front Immunol. 2021. PMID: 34349771 Free PMC article. Review.
-
Regnase-1-related endoribonucleases in health and immunological diseases.Immunol Rev. 2021 Nov;304(1):97-110. doi: 10.1111/imr.13023. Epub 2021 Sep 12. Immunol Rev. 2021. PMID: 34514623 Review.
-
RNA-binding protein-mediated post-transcriptional controls of gene expression: integration of molecular mechanisms at the 3' end of mRNAs?Biochem Pharmacol. 2014 Jun 15;89(4):431-40. doi: 10.1016/j.bcp.2014.04.003. Epub 2014 Apr 13. Biochem Pharmacol. 2014. PMID: 24735612
Cited by
-
Shaping Heterogeneity of Naive CD8+ T Cell Pools.Immune Netw. 2023 Feb 22;23(1):e2. doi: 10.4110/in.2023.23.e2. eCollection 2023 Feb. Immune Netw. 2023. PMID: 36911807 Free PMC article. Review.
-
Paradoxical imbalance between activated lymphocyte protein synthesis capacity and rapid division rate.Elife. 2024 Mar 21;12:RP89015. doi: 10.7554/eLife.89015. Elife. 2024. PMID: 38512721 Free PMC article.
-
Peripheral Blood CD8+ T-Lymphocyte Immune Response in Benign and Subpopulations of Breast Cancer Patients.Int J Mol Sci. 2024 Jun 11;25(12):6423. doi: 10.3390/ijms25126423. Int J Mol Sci. 2024. PMID: 38928129 Free PMC article.
References
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

