Calculation of centralities in protein kinase A
- PMID: 36375071
- PMCID: PMC9704751
- DOI: 10.1073/pnas.2215420119
Calculation of centralities in protein kinase A
Abstract
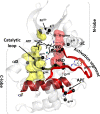
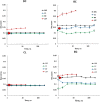
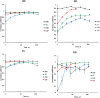
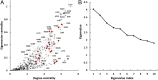
Topological analysis of protein residue networks (PRNs) is a common method that can help to understand the roles of individual residues. Here, we used protein kinase A as a study object and asked what already known functionally important residues can be detected by network analysis. Along several traditional approaches to weight edges in PRNs we used local spatial pattern (LSP) alignment that assigns high weights to edges only if CαCβ vectors for the corresponding residues retain their mutual positions and orientation. Our results show that even short molecular dynamic simulations of 10 to 20 ns can give convergent values for betweenness and degree centralities calculated from the LSP-based PRNs. Using these centralities, we were able to clearly distinguish a group of residues that are highly conserved in protein kinases and play important functional and regulatory roles. In comparison, traditional methods based on cross-correlation and linear mutual information were much less efficient for this particular task. These results call for reevaluation of the current methods to generate PRNs.
Keywords: allostery; network analysis; protein kinases.
Conflict of interest statement
Competing interest statement: P.C.A. has been employed by Eli Lilly after the completion of the work.
Figures
Similar articles
-
Gauging Dynamics-driven Allostery Using a New Computational Tool: A CAP Case Study.J Mol Biol. 2024 Jan 15;436(2):168395. doi: 10.1016/j.jmb.2023.168395. Epub 2023 Dec 13. J Mol Biol. 2024. PMID: 38097109 Free PMC article.
-
gRINN: a tool for calculation of residue interaction energies and protein energy network analysis of molecular dynamics simulations.Nucleic Acids Res. 2018 Jul 2;46(W1):W554-W562. doi: 10.1093/nar/gky381. Nucleic Acids Res. 2018. PMID: 29800260 Free PMC article.
-
The adaptive nature of protein residue networks.Proteins. 2017 May;85(5):917-923. doi: 10.1002/prot.25261. Epub 2017 Feb 16. Proteins. 2017. PMID: 28168745
-
A historical overview of protein kinases and their targeted small molecule inhibitors.Pharmacol Res. 2015 Oct;100:1-23. doi: 10.1016/j.phrs.2015.07.010. Epub 2015 Jul 21. Pharmacol Res. 2015. PMID: 26207888 Review.
-
From structure to the dynamic regulation of a molecular switch: A journey over 3 decades.J Biol Chem. 2021 Jan-Jun;296:100746. doi: 10.1016/j.jbc.2021.100746. Epub 2021 May 3. J Biol Chem. 2021. PMID: 33957122 Free PMC article. Review.
Cited by
-
Gauging Dynamics-driven Allostery Using a New Computational Tool: A CAP Case Study.J Mol Biol. 2024 Jan 15;436(2):168395. doi: 10.1016/j.jmb.2023.168395. Epub 2023 Dec 13. J Mol Biol. 2024. PMID: 38097109 Free PMC article.
-
Activation of hepatic adenosine A1 receptor ameliorates MASH via inhibiting SREBPs maturation.Cell Rep Med. 2024 Mar 19;5(3):101477. doi: 10.1016/j.xcrm.2024.101477. Cell Rep Med. 2024. PMID: 38508143 Free PMC article.
-
Temperature-dependent hydrogen deuterium exchange shows impact of analog binding on adenosine deaminase flexibility but not embedded thermal networks.J Biol Chem. 2022 Sep;298(9):102350. doi: 10.1016/j.jbc.2022.102350. Epub 2022 Aug 4. J Biol Chem. 2022. PMID: 35933011 Free PMC article.
-
Single-residue mutation in protein kinase C toggles between cancer and neurodegeneration.Biochem J. 2023 Aug 30;480(16):1299-1316. doi: 10.1042/BCJ20220397. Biochem J. 2023. PMID: 37551632 Free PMC article.
-
The "violin model": Looking at community networks for dynamic allostery.J Chem Phys. 2023 Feb 28;158(8):081001. doi: 10.1063/5.0138175. J Chem Phys. 2023. PMID: 36859094 Free PMC article. Review.
References
-
- Boccaletti S., Latora V., Moreno Y., Chavez M., Hwang D. U., Complex networks: Structure and dynamics. Phys. Rep. 424, 175–308 (2006).
-
- Christley R. M., et al. , Infection in social networks: Using network analysis to identify high-risk individuals. Am. J. Epidemiol. 162, 1024–1031 (2005). - PubMed
-
- Reggiani A., Nijkamp P., Lanzi D., Transport resilience and vulnerability: The role of connectivity. Transp. Res. Part A Policy Pract. 8, 4–15 (2015).
-
- Bavelas A., Communication patterns in task-oriented groups. J. Acoust. Soc. Am. 22, 723–730 (1950).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

