Longer Duration of Active Oil Biosynthesis during Seed Development Is Crucial for High Oil Yield-Lessons from Genome-Wide In Silico Mining and RNA-Seq Validation in Sesame
- PMID: 36365434
- PMCID: PMC9657858
- DOI: 10.3390/plants11212980
Longer Duration of Active Oil Biosynthesis during Seed Development Is Crucial for High Oil Yield-Lessons from Genome-Wide In Silico Mining and RNA-Seq Validation in Sesame
Abstract
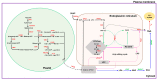
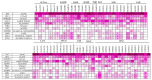
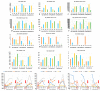
Sesame, one of the ancient oil crops, is an important oilseed due to its nutritionally rich seeds with high protein content. Genomic scale information for sesame has become available in the public databases in recent years. The genes and their families involved in oil biosynthesis in sesame are less studied than in other oilseed crops. Therefore, we retrieved a total of 69 genes and their translated amino acid sequences, associated with gene families linked to the oil biosynthetic pathway. Genome-wide in silico mining helped identify key regulatory genes for oil biosynthesis, though the findings require functional validation. Comparing sequences of the SiSAD (stearoyl-acyl carrier protein (ACP)-desaturase) coding genes with known SADs helped identify two SiSAD family members that may be palmitoyl-ACP-specific. Based on homology with lysophosphatidic acid acyltransferase (LPAAT) sequences, an uncharacterized gene has been identified as SiLPAAT1. Identified key regulatory genes associated with high oil content were also validated using publicly available transcriptome datasets of genotypes contrasting for oil content at different developmental stages. Our study provides evidence that a longer duration of active oil biosynthesis is crucial for high oil accumulation during seed development. This underscores the importance of early onset of oil biosynthesis in developing seeds. Up-regulating, identified key regulatory genes of oil biosynthesis during early onset of seed development, should help increase oil yields.
Keywords: ACCase; DGAT; FAD; KAS; Kennedy pathway; LACS; LPAAT; SAD; early onset; longer duration; oil biosynthesis; sesame.
Conflict of interest statement
All authors declare that there are no direct or indirect, including financial and non-financial, competing interests, with reference to this article.
Figures

Similar articles
-
A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality.BMC Genomics. 2017 Mar 1;18(1):218. doi: 10.1186/s12864-017-3594-9. BMC Genomics. 2017. PMID: 28249560 Free PMC article.
-
Integrated lipidomic and transcriptomic analyses reveal the mechanism of lipid biosynthesis and accumulation during seed development in sesame.Front Plant Sci. 2023 Jun 22;14:1211040. doi: 10.3389/fpls.2023.1211040. eCollection 2023. Front Plant Sci. 2023. PMID: 37426956 Free PMC article.
-
Gene expression profiles that shape high and low oil content sesames.BMC Genet. 2019 May 16;20(1):45. doi: 10.1186/s12863-019-0747-7. BMC Genet. 2019. PMID: 31096908 Free PMC article.
-
Comparative analysis of transcriptome in oil biosynthesis between seeds and non-seed tissues of Symplocos paniculata fruit.Front Plant Sci. 2024 Oct 2;15:1441602. doi: 10.3389/fpls.2024.1441602. eCollection 2024. Front Plant Sci. 2024. PMID: 39416484 Free PMC article.
-
Analysis of expression sequence tags from a full-length-enriched cDNA library of developing sesame seeds (Sesamum indicum).BMC Plant Biol. 2011 Dec 24;11:180. doi: 10.1186/1471-2229-11-180. BMC Plant Biol. 2011. PMID: 22195973 Free PMC article.
Cited by
-
Evaluation and validation of suitable reference genes for quantitative real-time PCR analysis in lotus (Nelumbo nucifera Gaertn.).Sci Rep. 2024 May 13;14(1):10857. doi: 10.1038/s41598-024-61806-9. Sci Rep. 2024. PMID: 38740848 Free PMC article.
-
Advances in Single-Cell Sequencing Technology and Its Application in Poultry Science.Genes (Basel). 2022 Nov 25;13(12):2211. doi: 10.3390/genes13122211. Genes (Basel). 2022. PMID: 36553479 Free PMC article. Review.
-
Genome-wide characterization of the soybean DOMAIN OF UNKNOWN FUNCTION 679 membrane protein gene family highlights their potential involvement in growth and stress response.Front Plant Sci. 2023 Sep 8;14:1216082. doi: 10.3389/fpls.2023.1216082. eCollection 2023. Front Plant Sci. 2023. PMID: 37745995 Free PMC article.
References
-
- Bruinsma J. World Agriculture: Towards 2015/2030-An FAO Perspective. Taylor and Francis; London, UK: 2017. p. 444.
-
- Banaś W., Sanchez Garcia A., Banaś A., Stymne S. Activities of acyl-CoA: Diacylglycerol acyltransferase (DGAT) and phospholipid: Diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds. Planta. 2013;237:1627–1636. doi: 10.1007/s00425-013-1870-8. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

