Development and Applications of CRISPR/Cas9-Based Genome Editing in Lactobacillus
- PMID: 36361647
- PMCID: PMC9656040
- DOI: 10.3390/ijms232112852
Development and Applications of CRISPR/Cas9-Based Genome Editing in Lactobacillus
Abstract
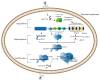
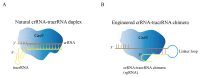
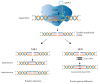
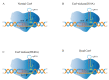
Lactobacillus, a genus of lactic acid bacteria, plays a crucial function in food production preservation, and probiotics. It is particularly important to develop new Lactobacillus strains with superior performance by gene editing. Currently, the identification of its functional genes and the mining of excellent functional genes mainly rely on the traditional gene homologous recombination technology. CRISPR/Cas9-based genome editing is a rapidly developing technology in recent years. It has been widely applied in mammalian cells, plants, yeast, and other eukaryotes, but less in prokaryotes, especially Lactobacillus. Compared with the traditional strain improvement methods, CRISPR/Cas9-based genome editing can greatly improve the accuracy of Lactobacillus target sites and achieve traceless genome modification. The strains obtained by this technology may even be more efficient than the traditional random mutation methods. This review examines the application and current issues of CRISPR/Cas9-based genome editing in Lactobacillus, as well as the development trend of CRISPR/Cas9-based genome editing in Lactobacillus. In addition, the fundamental mechanisms of CRISPR/Cas9-based genome editing are also presented and summarized.
Keywords: CRISPR/Cas9; Cas9 protein; Lactobacillus; genome editing.
Conflict of interest statement
The authors here declare that they have no known competing financial interests or personal relationships that could have influenced this research.
Figures
Similar articles
-
Portable CRISPR-Cas9N System for Flexible Genome Engineering in Lactobacillus acidophilus, Lactobacillus gasseri, and Lactobacillus paracasei.Appl Environ Microbiol. 2021 Feb 26;87(6):e02669-20. doi: 10.1128/AEM.02669-20. Print 2021 Feb 26. Appl Environ Microbiol. 2021. PMID: 33397707 Free PMC article.
-
Development of a RecE/T-Assisted CRISPR-Cas9 Toolbox for Lactobacillus.Biotechnol J. 2019 Jul;14(7):e1800690. doi: 10.1002/biot.201800690. Epub 2019 May 20. Biotechnol J. 2019. PMID: 30927506
-
CRISPR/Cas9-Assisted Seamless Genome Editing in Lactobacillus plantarum and Its Application in N-Acetylglucosamine Production.Appl Environ Microbiol. 2019 Oct 16;85(21):e01367-19. doi: 10.1128/AEM.01367-19. Print 2019 Nov 1. Appl Environ Microbiol. 2019. PMID: 31444197 Free PMC article.
-
[Application of gene editing technology in Escherichia coli].Sheng Wu Gong Cheng Xue Bao. 2022 Apr 25;38(4):1446-1461. doi: 10.13345/j.cjb.210680. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 35470618 Review. Chinese.
-
Can genetic engineering-based methods for gene function identification be eclipsed by genome editing in plants? A comparison of methodologies.Mol Genet Genomics. 2021 May;296(3):485-500. doi: 10.1007/s00438-021-01769-y. Epub 2021 Mar 9. Mol Genet Genomics. 2021. PMID: 33751237 Review.
Cited by
-
The chicken chorioallantoic membrane model for isolation of CRISPR/cas9-based HSV-1 mutant expressing tumor suppressor p53.PLoS One. 2023 Oct 20;18(10):e0286231. doi: 10.1371/journal.pone.0286231. eCollection 2023. PLoS One. 2023. PMID: 37862369 Free PMC article.
-
The arsenic bioremediation using genetically engineered microbial strains on aquatic environments: An updated overview.Heliyon. 2024 Aug 22;10(17):e36314. doi: 10.1016/j.heliyon.2024.e36314. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39286167 Free PMC article. Review.
References
-
- Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. - DOI - PubMed
-
- Leuschner R.G.K., Robinson T.P., Hugas M., Cocconcelli P.S., Richard-Forget F., Klein G., Licht T.R., Nguyen-The C., Querol A., Richardson M., et al. Qualified presumption of safety (QPS): A generic risk assessment approach for biological agents notified to the European Food Safety Authority (EFSA) Trends Food Sci. Technol. 2010;21:425–435. doi: 10.1016/j.tifs.2010.07.003. - DOI
-
- Rocchetti M.T., Russo P., Spano G., De Santis L., Iarusso I., De Simone N., Brahimi S., Fiocco D., Capozzi V. Exploring the Probiotic Potential of Dairy Industrial-Relevant Lactobacilli. Appl. Sci. 2022;12:4989. doi: 10.3390/app12104989. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

