Novel protein complexes containing autophagy and UPS components regulate proteasome-dependent PARK2 recruitment onto mitochondria and PARK2-PARK6 activity during mitophagy
- PMID: 36357363
- PMCID: PMC9649694
- DOI: 10.1038/s41419-022-05339-x
Novel protein complexes containing autophagy and UPS components regulate proteasome-dependent PARK2 recruitment onto mitochondria and PARK2-PARK6 activity during mitophagy
Abstract
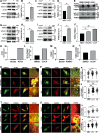
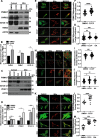
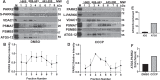
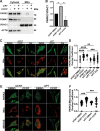
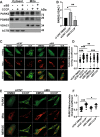
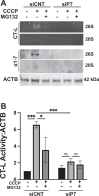
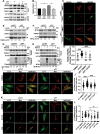
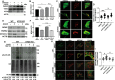
Autophagy is an evolutionarily conserved eukaryotic cellular mechanism through which cytosolic fragments, misfolded/aggregated proteins and organelles are degraded and recycled. Priming of mitochondria through ubiquitylation is required for the clearance the organelle by autophagy (mitophagy). Familial Parkinson's Disease-related proteins, including the E3-ligase PARK2 (PARKIN) and the serine/threonine kinase PARK6 (PINK1) control these ubiquitylation reactions and contribute to the regulation of mitophagy. Here we describe, novel protein complexes containing autophagy protein ATG5 and ubiquitin-proteasome system (UPS) components. We discovered that ATG5 interacts with PSMA7 and PARK2 upon mitochondrial stress. Results suggest that all three proteins translocate mitochondria and involve in protein complexes containing autophagy, UPS and mitophagy proteins. Interestingly, PARK2 and ATG5 recruitment onto mitochondria requires proteasome components PSMA7 and PSMB5. Strikingly, we discovered that subunit of 20 S proteasome, PSMA7, is required for the progression of PARK2-PARK6-mediated mitophagy and the proteasome activity following mitochondrial stress. Our results demonstrate direct, dynamic and functional interactions between autophagy and UPS components that contribute to the regulation of mitophagy.
© 2022. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare. All co-authors have seen and agree with the contents of the manuscript and there is no financial interest to report. The authors declare no competing interests.
Figures
Similar articles
-
Deubiquitinating enzymes regulate PARK2-mediated mitophagy.Autophagy. 2015 Apr 3;11(4):595-606. doi: 10.1080/15548627.2015.1034408. Autophagy. 2015. PMID: 25915564 Free PMC article.
-
PARK2/Parkin-mediated mitochondrial clearance contributes to proteasome activation during slow-twitch muscle atrophy via NFE2L1 nuclear translocation.Autophagy. 2014 Apr;10(4):631-41. doi: 10.4161/auto.27785. Epub 2014 Jan 21. Autophagy. 2014. PMID: 24451648 Free PMC article.
-
Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy.Hum Mol Genet. 2011 May 1;20(9):1726-37. doi: 10.1093/hmg/ddr048. Epub 2011 Feb 4. Hum Mol Genet. 2011. PMID: 21296869 Free PMC article.
-
N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review.
-
Twenty years since the discovery of the parkin gene.J Neural Transm (Vienna). 2017 Sep;124(9):1037-1054. doi: 10.1007/s00702-017-1742-7. Epub 2017 Jun 15. J Neural Transm (Vienna). 2017. PMID: 28620835 Review.
Cited by
-
Multiple roles of mitochondrial autophagy receptor FUNDC1 in mitochondrial events and kidney disease.Front Cell Dev Biol. 2024 Oct 9;12:1453365. doi: 10.3389/fcell.2024.1453365. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39445333 Free PMC article. Review.
-
Identification of Autophagy-Related Genes in Patients with Acute Spinal Cord Injury and Analysis of Potential Therapeutic Targets.Mol Neurobiol. 2025 Mar;62(3):2674-2694. doi: 10.1007/s12035-024-04431-x. Epub 2024 Aug 16. Mol Neurobiol. 2025. PMID: 39150631
-
Autophagy protects mitochondrial health in heart failure.Heart Fail Rev. 2024 Jan;29(1):113-123. doi: 10.1007/s10741-023-10354-x. Epub 2023 Oct 12. Heart Fail Rev. 2024. PMID: 37823952 Review.
-
Interactome of Arabidopsis ATG5 Suggests Functions beyond Autophagy.Int J Mol Sci. 2023 Aug 1;24(15):12300. doi: 10.3390/ijms241512300. Int J Mol Sci. 2023. PMID: 37569688 Free PMC article.
-
Osteogenesis imperfecta type 10 and the cellular scaffolds underlying common immunological diseases.Genes Immun. 2024 Aug;25(4):265-276. doi: 10.1038/s41435-024-00277-4. Epub 2024 May 29. Genes Immun. 2024. PMID: 38811682 Review.
References
-
- Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–73. - PubMed
-
- Walker S, Chandra P, Manifava M, Axe E, Ktistakis NT. Making autophagosomes: Localized synthesis of phosphatidylinositol 3-phosphate holds the clue. Autophagy. 2008;4:1093–6. - PubMed
-
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. - PubMed
-
- Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–41. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

