Silica nanoparticles induce ovarian granulosa cell apoptosis via activation of the PERK-ATF4-CHOP-ERO1α pathway-mediated IP3R1-dependent calcium mobilization
- PMID: 36346508
- PMCID: PMC10604358
- DOI: 10.1007/s10565-022-09776-4
Silica nanoparticles induce ovarian granulosa cell apoptosis via activation of the PERK-ATF4-CHOP-ERO1α pathway-mediated IP3R1-dependent calcium mobilization
Abstract
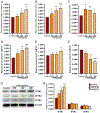
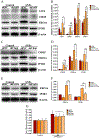
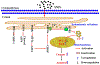
Ambient particulate matters (PMs) have adverse effects in human and animal female reproductive health. Silica nanoparticles (SNPs), as a major component of PMs, can induce follicular atresia via the promotion of ovarian granulosa cell apoptosis. However, the molecular mechanisms of apoptosis induced by SNPs are not very clear. This work focuses on revealing the mechanisms of ER stress on SNP-induced apoptosis. Our results showed that spherical Stöber SNPs (110 nm, 25.0 mg/kg b.w.) induced follicular atresia via the promotion of granulosa cell apoptosis by intratracheal instillation in vivo; meanwhile, SNPs decreased the viability and increase apoptosis in granulosa cells in vitro. SNPs were taken up and accumulated in the vesicles of granulosa cells. Additionally, our results found that SNPs increased calcium ion (Ca2+) concentration in granulosa cell cytoplasm. Furthermore, SNPs activated ER stress via an increase in the PERK and ATF6 pathway-related protein levels and IP3R1-dependent calcium mobilization via an increase in IP3R1 level. In addition, 4-PBA restored IP3R1-dependent calcium mobilization and decreased apoptosis via the inhibition of ER stress. The ATF4-C/EBP homologous protein (CHOP)-ER oxidoreductase 1 alpha (ERO1α) pathway regulated SNP-induced IP3R1-dependent calcium mobilization and cell apoptosis via ATF4, CHOP, and ERO1α depletion in ovarian granulosa cells. Herein, we demonstrate that ER stress cooperated in SNP-induced ovarian toxicity via activation of IP3R1-mediated calcium mobilization, leading to apoptosis, in which the PERK-ATF4-CHOP-ERO1α pathway plays an essential role in ovarian granulosa cells.
Keywords: Apoptosis; Calcium mobilization; ER stress; Follicular atresia; Ovarian granulosa cells; Silica nanoparticles.
© 2022. The Author(s), under exclusive licence to Springer Nature B.V.
Conflict of interest statement
Figures









Similar articles
-
Silica Nanoparticles Promote Apoptosis in Ovarian Granulosa Cells via Autophagy Dysfunction.Int J Mol Sci. 2023 Mar 8;24(6):5189. doi: 10.3390/ijms24065189. Int J Mol Sci. 2023. PMID: 36982262 Free PMC article.
-
Endoplasmic reticulum stress is involved in granulosa cell apoptosis during follicular atresia in goat ovaries.Mol Reprod Dev. 2012 Jun;79(6):423-32. doi: 10.1002/mrd.22045. Epub 2012 May 4. Mol Reprod Dev. 2012. PMID: 22532439
-
Protegrin-1 inhibits porcine ovarian granulosa cell apoptosis from H2O2-induced oxidative stress via the PERK/eIF2α/CHOP signaling pathway in vitro.Theriogenology. 2022 Feb;179:117-127. doi: 10.1016/j.theriogenology.2021.11.022. Epub 2021 Nov 29. Theriogenology. 2022. PMID: 34864562
-
The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review.
-
Advances in the Effects of Heat Stress on Ovarian Granulosa Cells: Unveiling Novel Ferroptosis Pathways.Vet Sci. 2024 Oct 1;11(10):464. doi: 10.3390/vetsci11100464. Vet Sci. 2024. PMID: 39453056 Free PMC article. Review.
Cited by
-
Taurine Protects against Silica Nanoparticle-Induced Apoptosis and Inflammatory Response via Inhibition of Oxidative Stress in Porcine Ovarian Granulosa Cells.Animals (Basel). 2024 Oct 14;14(20):2959. doi: 10.3390/ani14202959. Animals (Basel). 2024. PMID: 39457890 Free PMC article.
-
Silica Nanoparticles Promote Apoptosis in Ovarian Granulosa Cells via Autophagy Dysfunction.Int J Mol Sci. 2023 Mar 8;24(6):5189. doi: 10.3390/ijms24065189. Int J Mol Sci. 2023. PMID: 36982262 Free PMC article.
-
Calcium signaling in oocyte quality and functionality and its application.Front Endocrinol (Lausanne). 2024 Aug 16;15:1411000. doi: 10.3389/fendo.2024.1411000. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39220364 Free PMC article. Review.
References
-
- Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J Biol Chem. 2004;279:5288–97. - PubMed
-
- Bitar A, Ahmad NM, Fessi H, Elaissari A. Silica-based nanoparticles for biomedical applications. Drug Discov Today. 2012;17:1147–54. - PubMed
-
- Che L, Yao H, Yang CL, Guo NJ, Huang J, Wu ZL, et al. Cyclooxygenase-2 modulates ER-mitochondria crosstalk to mediate superparamagnetic iron oxide nanoparticles induced hepatotoxicity: an in vitro and in vivo study. Nanotoxicology. 2020;14:162–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

