Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops
- PMID: 36335136
- PMCID: PMC9637178
- DOI: 10.1038/s41467-022-34276-8
Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops
Abstract
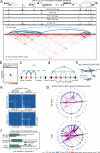
The three-dimensional genomic structure plays a critical role in gene expression, cellular differentiation, and pathological conditions. It is pivotal to elucidate fine-scale chromatin architectures, especially interactions of regulatory elements, to understand the temporospatial regulation of gene expression. In this study, we report Hi-TrAC as a proximity ligation-free, robust, and sensitive technique to profile genome-wide chromatin interactions at high-resolution among regulatory elements. Hi-TrAC detects chromatin looping among accessible regions at single nucleosome resolution. With almost half-million identified loops, we reveal a comprehensive interaction network of regulatory elements across the genome. After integrating chromatin binding profiles of transcription factors, we discover that cohesin complex and CTCF are responsible for organizing long-range chromatin loops, related to domain formation; whereas ZNF143 and HCFC1 are involved in structuring short-range chromatin loops between regulatory elements, which directly regulate gene expression. Thus, we introduce a methodology to identify a delicate and comprehensive network of cis-regulatory elements, revealing the complexity and a division of labor of transcription factors in organizing chromatin loops for genome organization and gene expression.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs.BMC Genomics. 2019 Oct 25;20(1):777. doi: 10.1186/s12864-019-6088-0. BMC Genomics. 2019. PMID: 31653198 Free PMC article.
-
Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins.EMBO J. 2017 Dec 15;36(24):3573-3599. doi: 10.15252/embj.201798004. Epub 2017 Dec 7. EMBO J. 2017. PMID: 29217591 Free PMC article.
-
Analysis of long-range chromatin contacts, compartments and looping between mouse embryonic stem cells, lens epithelium and lens fibers.Epigenetics Chromatin. 2024 Apr 20;17(1):10. doi: 10.1186/s13072-024-00533-x. Epigenetics Chromatin. 2024. PMID: 38643244 Free PMC article.
-
Architectural proteins for the formation and maintenance of the 3D genome.Sci China Life Sci. 2020 Jun;63(6):795-810. doi: 10.1007/s11427-019-1613-3. Epub 2020 Apr 2. Sci China Life Sci. 2020. PMID: 32249389 Review.
-
A tour of 3D genome with a focus on CTCF.Semin Cell Dev Biol. 2019 Jun;90:4-11. doi: 10.1016/j.semcdb.2018.07.020. Epub 2018 Jul 23. Semin Cell Dev Biol. 2019. PMID: 30031214 Review.
Cited by
-
ZNF143 binds DNA and stimulates transcripstion initiation to activate and repress direct target genes.bioRxiv [Preprint]. 2024 May 15:2024.05.13.594008. doi: 10.1101/2024.05.13.594008. bioRxiv. 2024. Update in: Nucleic Acids Res. 2025 Jan 11;53(2):gkae1182. doi: 10.1093/nar/gkae1182. PMID: 38798607 Free PMC article. Updated. Preprint.
-
Systems immunology approaches to study T cells in health and disease.NPJ Syst Biol Appl. 2024 Oct 9;10(1):117. doi: 10.1038/s41540-024-00446-1. NPJ Syst Biol Appl. 2024. PMID: 39384819 Free PMC article. Review.
-
Function and Evolution of the Loop Extrusion Machinery in Animals.Int J Mol Sci. 2023 Mar 6;24(5):5017. doi: 10.3390/ijms24055017. Int J Mol Sci. 2023. PMID: 36902449 Free PMC article. Review.
-
Loop Catalog: a comprehensive HiChIP database of human and mouse samples.bioRxiv [Preprint]. 2025 Mar 6:2024.04.26.591349. doi: 10.1101/2024.04.26.591349. bioRxiv. 2025. PMID: 38746164 Free PMC article. Preprint.
-
DeepCORE: An interpretable multi-view deep neural network model to detect co-operative regulatory elements.Comput Struct Biotechnol J. 2023 Dec 29;23:679-687. doi: 10.1016/j.csbj.2023.12.044. eCollection 2024 Dec. Comput Struct Biotechnol J. 2023. PMID: 38292477 Free PMC article.
References
-
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016;17:661–678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

