A proto-telomere is elongated by telomerase in a shelterin-dependent manner in quiescent fission yeast cells
- PMID: 36330920
- PMCID: PMC9723628
- DOI: 10.1093/nar/gkac986
A proto-telomere is elongated by telomerase in a shelterin-dependent manner in quiescent fission yeast cells
Abstract
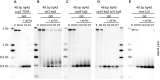
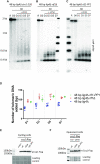
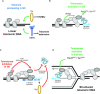
Telomere elongation is coupled with genome replication, raising the question of the repair of short telomeres in post-mitotic cells. We investigated the fate of a telomere-repeat capped end that mimics a single short telomere in quiescent fission yeast cells. We show that telomerase is able to elongate this single short telomere during quiescence despite the binding of Ku to the proto-telomere. While Taz1 and Rap1 repress telomerase in vegetative cells, both shelterin proteins are required for efficient telomere extension in quiescent cells, underscoring a distinct mode of telomerase control. We further show that Rad3ATR and Tel1ATM are redundantly required for telomere elongation in quiescence through the phosphorylation of Ccq1 and that Rif1 and its associated-PP1 phosphatases negatively regulate telomerase activity by opposing Ccq1 phosphorylation. The distinct mode of telomerase regulation in quiescent fission yeast cells may be relevant to that in human stem and progenitor cells.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Tpz1-Ccq1 and Tpz1-Poz1 interactions within fission yeast shelterin modulate Ccq1 Thr93 phosphorylation and telomerase recruitment.PLoS Genet. 2014 Oct 16;10(10):e1004708. doi: 10.1371/journal.pgen.1004708. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25330395 Free PMC article.
-
Fission yeast shelterin regulates DNA polymerases and Rad3(ATR) kinase to limit telomere extension.PLoS Genet. 2013 Nov;9(11):e1003936. doi: 10.1371/journal.pgen.1003936. Epub 2013 Nov 7. PLoS Genet. 2013. PMID: 24244195 Free PMC article.
-
Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.PLoS Genet. 2022 Jul 18;18(7):e1010308. doi: 10.1371/journal.pgen.1010308. eCollection 2022 Jul. PLoS Genet. 2022. PMID: 35849625 Free PMC article.
-
CST meets shelterin to keep telomeres in check.Mol Cell. 2010 Sep 10;39(5):665-76. doi: 10.1016/j.molcel.2010.08.024. Mol Cell. 2010. PMID: 20832719 Review.
-
Finding the end: recruitment of telomerase to telomeres.Nat Rev Mol Cell Biol. 2013 Feb;14(2):69-82. doi: 10.1038/nrm3505. Epub 2013 Jan 9. Nat Rev Mol Cell Biol. 2013. PMID: 23299958 Free PMC article. Review.
Cited by
-
A Rapidly Inducible DNA Double-Strand Break to Monitor Telomere Formation, DNA Repair, and Checkpoint Activation.Methods Mol Biol. 2025;2862:209-221. doi: 10.1007/978-1-0716-4168-2_15. Methods Mol Biol. 2025. PMID: 39527203
References
-
- Lingner J., Cooper J., Cech T.. Telomerase and DNA end replication: no longer a lagging strand problem?. Science. 1995; 269:1533–1534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

