Loss of miR-637 promotes cancer cell stemness via WASH/IL-8 pathway and serves as a novel prognostic marker in esophageal squamous cell carcinoma
- PMID: 36329557
- PMCID: PMC9635169
- DOI: 10.1186/s40364-022-00424-x
Loss of miR-637 promotes cancer cell stemness via WASH/IL-8 pathway and serves as a novel prognostic marker in esophageal squamous cell carcinoma
Abstract
Background: Esophageal carcinoma is the highly lethal cancer in the world, predominantly in some areas of East Asia. We previously reported that overexpression of cytoskeleton regulator Wiskott-Aldrich syndrome protein and SCAR Homolog (WASH) associates with poor prognosis of patients with esophageal squamous cell carcinoma (ESCC). However, the molecular mechanism and clinical significance involved in WASH overexpression have not been fully elucidated.
Methods: Bioinformatics analysis and luciferase reporter assay were used to predict and validate miR-637 as a regulator of WASH in ESCC cell lines. qRT-PCR, Western blotting and ELISA assays were performed to examine RNA expression and protein levels, respectively. Next, the biological functions of miR-637 were explored by tumor sphere formation assay in vitro and nude mouse tumor xenograft in vivo. Finally, we evaluated the association of miR-637 levels with clinical features in ESCC patients.
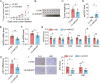
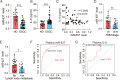
Results: We identified miR-637 as a WASH-targeting miRNA. miR-637 mimic strongly attenuated the downstream IL-8 production and tumor sphere formation in esophageal cancer cells, whereas miR-637 inhibitor displayed an opposite effect. IL-8 could facilitate stem-like properties and partially rescue the phenotypes induced by miR-637 mimic. Furthermore, miR-637 inhibitor dramatically promoted IL-8 expression and cancer stemness properties in a WASH-dependent manner. Ectopic expression of miR-637 also inhibited tumor growth in a mouse model. Clinically, low expression of miR-637 was observed in tumor tissues and the low expression levels of miR-637 were correlated with poor survival of ESCC patients. In particular, plasma miR-637 could be used as a noninvasive biomarker for ESCC patients.
Conclusions: These results implicate the potential application of miR-637 for diagnosis and prognosis of esophageal cancer.
Keywords: Cancer stem cells; Esophageal squamous cell carcinoma; Interleukin-8; WASH; miR-637.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
WASH overexpression enhances cancer stem cell properties and correlates with poor prognosis of esophageal carcinoma.Cancer Sci. 2017 Dec;108(12):2358-2365. doi: 10.1111/cas.13400. Epub 2017 Sep 26. Cancer Sci. 2017. PMID: 28914471 Free PMC article.
-
Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC).J Exp Clin Cancer Res. 2018 Dec 4;37(1):301. doi: 10.1186/s13046-018-0966-1. J Exp Clin Cancer Res. 2018. PMID: 30514328 Free PMC article.
-
Circular RNA circNTRK2 facilitates the progression of esophageal squamous cell carcinoma through up-regulating NRIP1 expression via miR-140-3p.J Exp Clin Cancer Res. 2020 Jul 11;39(1):133. doi: 10.1186/s13046-020-01640-9. J Exp Clin Cancer Res. 2020. PMID: 32653032 Free PMC article.
-
Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma.J Exp Clin Cancer Res. 2017 Nov 16;36(1):161. doi: 10.1186/s13046-017-0622-1. J Exp Clin Cancer Res. 2017. PMID: 29145896 Free PMC article.
-
microRNA-messenger RNA regulatory network of esophageal squamous cell carcinoma and the identification of miR-1 as a biomarker of patient survival.J Cell Biochem. 2019 Aug;120(8):12259-12272. doi: 10.1002/jcb.28166. Epub 2019 Apr 24. J Cell Biochem. 2019. PMID: 31017699
Cited by
-
Human esophageal cancer stem-like cells escape the cytotoxicity of natural killer cells via down-regulation of ULBP-1.J Transl Med. 2024 Aug 5;22(1):737. doi: 10.1186/s12967-024-05549-1. J Transl Med. 2024. PMID: 39103915 Free PMC article.
References
LinkOut - more resources
Full Text Sources

