Arginine ADP-Ribosylation: Chemical Synthesis of Post-Translationally Modified Ubiquitin Proteins
- PMID: 36318515
- PMCID: PMC9673145
- DOI: 10.1021/jacs.2c06249
Arginine ADP-Ribosylation: Chemical Synthesis of Post-Translationally Modified Ubiquitin Proteins
Abstract
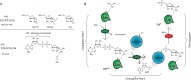
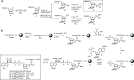
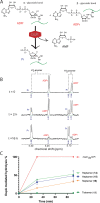
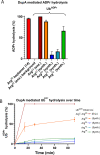
We describe the development and optimization of a methodology to prepare peptides and proteins modified on the arginine residue with an adenosine-di-phosphate-ribosyl (ADPr) group. Our method comprises reacting an ornithine containing polypeptide on-resin with an α-linked anomeric isothiourea N-riboside, ensuing installment of a phosphomonoester at the 5'-hydroxyl of the ribosyl moiety followed by the conversion into the adenosine diphosphate. We use this method to obtain four regioisomers of ADP-ribosylated ubiquitin (UbADPr), each modified with an ADP-ribosyl residue on a different arginine position within the ubiquitin (Ub) protein (Arg42, Arg54, Arg72, and Arg74) as the first reported examples of fully synthetic arginine-linked ADPr-modified proteins. We show the chemically prepared Arg-linked UbADPr to be accepted and processed by Legionella enzymes and compare the entire suite of four Arg-linked UbADPr regioisomers in a variety of biochemical experiments, allowing us to profile the activity and selectivity of Legionella pneumophila ligase and hydrolase enzymes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
A General Approach Towards Triazole-Linked Adenosine Diphosphate Ribosylated Peptides and Proteins.Angew Chem Int Ed Engl. 2018 Feb 5;57(6):1659-1662. doi: 10.1002/anie.201710527. Epub 2018 Jan 8. Angew Chem Int Ed Engl. 2018. PMID: 29215186
-
Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches.J Am Soc Mass Spectrom. 2021 Jan 6;32(1):157-168. doi: 10.1021/jasms.0c00040. Epub 2020 Nov 3. J Am Soc Mass Spectrom. 2021. PMID: 33140951
-
Generating Protein-Linked and Protein-Free Mono-, Oligo-, and Poly(ADP-Ribose) In Vitro.Methods Mol Biol. 2018;1813:91-108. doi: 10.1007/978-1-4939-8588-3_7. Methods Mol Biol. 2018. PMID: 30097863 Free PMC article.
-
Chemical ADP-ribosylation: mono-ADPr-peptides and oligo-ADP-ribose.Org Biomol Chem. 2019 Jun 5;17(22):5460-5474. doi: 10.1039/c9ob00501c. Org Biomol Chem. 2019. PMID: 31112180 Review.
-
Chemical Tools to Study Protein ADP-Ribosylation.ACS Omega. 2020 Jan 22;5(4):1743-1751. doi: 10.1021/acsomega.9b03591. eCollection 2020 Feb 4. ACS Omega. 2020. PMID: 32039309 Free PMC article. Review.
Cited by
-
Reversal of tyrosine-linked ADP-ribosylation by ARH3 and PARG.J Biol Chem. 2024 Nov;300(11):107838. doi: 10.1016/j.jbc.2024.107838. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39342999 Free PMC article.
-
Capturing Legionella pneumophila effector enzymes using a ubiquitin derived photo-activatable probe.Front Mol Biosci. 2024 Jul 9;11:1422034. doi: 10.3389/fmolb.2024.1422034. eCollection 2024. Front Mol Biosci. 2024. PMID: 39044841 Free PMC article.
-
Legionella metaeffector MavL reverses ubiquitin ADP-ribosylation via a conserved arginine-specific macrodomain.Nat Commun. 2024 Mar 19;15(1):2452. doi: 10.1038/s41467-024-46649-2. Nat Commun. 2024. PMID: 38503748 Free PMC article.
-
PARP14 is a PARP with both ADP-ribosyl transferase and hydrolase activities.Sci Adv. 2023 Sep 15;9(37):eadi2687. doi: 10.1126/sciadv.adi2687. Epub 2023 Sep 13. Sci Adv. 2023. PMID: 37703374 Free PMC article.
-
PARPs and ADP-ribosylation: Deciphering the complexity with molecular tools.Mol Cell. 2023 May 18;83(10):1552-1572. doi: 10.1016/j.molcel.2023.04.009. Epub 2023 Apr 28. Mol Cell. 2023. PMID: 37119811 Free PMC article. Review.
References
-
- Ueda K.; Hayaishi O.; Oka J.; Komura H.; Nakanishi K.. ADP-Ribosylation of Proteins; Althaus F. R.; Hilz H.; Shall S., Eds.; Springer: Berlin, 1985; pp 159–166.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

