Arginase-2-specific cytotoxic T cells specifically recognize functional regulatory T cells
- PMID: 36316062
- PMCID: PMC9628693
- DOI: 10.1136/jitc-2022-005326
Arginase-2-specific cytotoxic T cells specifically recognize functional regulatory T cells
Abstract
Background: High expression of the metabolic enzyme arginase-2 (ARG2) by cancer cells, regulatory immune cells, or cells of the tumor stroma can reduce the availability of arginine (L-Arg) in the tumor microenvironment (TME). Depletion of L-Arg has detrimental consequences for T cells and leads to T-cell dysfunction and suppression of anticancer immune responses. Previous work from our group has demonstrated the presence of proinflammatory ARG2-specific CD4 T cells that inhibited tumor growth in murine models on activation with ARG2-derived peptides. In this study, we investigated the natural occurrence of ARG2-specific CD8 T cells in both healthy donors (HDs) and patients with cancer, along with their immunomodulatory capabilities in the context of the TME.
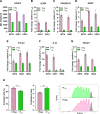
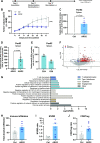
Materials and methods: A library of 15 major histocompatibility complex (MHC) class I-restricted ARG2-derived peptides were screened in HD peripheral blood mononuclear cells using interferon gamma (IFN-γ) ELISPOT. ARG2-specific CD8 T-cell responses were identified using intracellular cytokine staining and ARG2-specific CD8 T-cell cultures were established by enrichment and rapid expansion following in vitro peptide stimulation. The reactivity of the cultures toward ARG2-expressing cells, including cancer cell lines and activated regulatory T cells (Tregs), was assessed using IFN-γ ELISPOT and a chromium release assay. The Treg signature was validated based on proliferation suppression assays, flow cytometry and quantitative reverse transcription PCR (RT-qPCR). In addition, vaccinations with ARG2-derived epitopes were performed in the murine Pan02 tumor model, and induction of ARG2-specific T-cell responses was evaluated with IFN-γ ELISPOT. RNAseq and subsequent GO-term and ImmuCC analysis was performed on the tumor tissue.
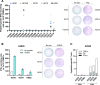
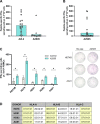
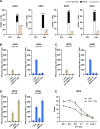
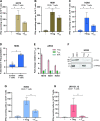
Results: We describe the existence of ARG2-specific CD8+ T cells and demonstrate these CD8+ T-cell responses in both HDs and patients with cancer. ARG2-specific T cells recognize and react to an ARG2-derived peptide presented in the context of HLA-B8 and exert their cytotoxic function against cancer cells with endogenous ARG2 expression. We demonstrate that ARG2-specific T cells can specifically recognize and react to activated Tregs with high ARG2 expression. Finally, we observe tumor growth suppression and antitumorigenic immunomodulation following ARG2 vaccination in an in vivo setting.
Conclusion: These findings highlight the ability of ARG2-specific T cells to modulate the immunosuppressive TME and suggest that ARG2-based immunomodulatory vaccines may be an interesting option for cancer immunotherapy.
Keywords: Arginase; Immunomodulation; Tumor Microenvironment; Vaccination.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: MHA is named as an inventor on various patent applications relating to the therapeutic use of arginase peptides, including ARG2 peptides, for vaccination. These patent applications have been transferred to the company IO Biotech ApS whose purpose is to develop immunomodulatory vaccines for cancer treatment. MHA is cofounder, shareholder and scientific advisor of IO Biotech ApS. IMS is cofounder, shareholder and clinical advisor of IO Biotech ApS. The remaining authors declare no conflicts of interest.
Figures
Similar articles
-
The metabolic enzyme arginase-2 is a potential target for novel immune modulatory vaccines.Oncoimmunology. 2020 Jun 1;9(1):1771142. doi: 10.1080/2162402X.2020.1771142. Oncoimmunology. 2020. PMID: 32923127 Free PMC article.
-
Identification of MHC class II restricted T-cell-mediated reactivity against MHC class I binding Mycobacterium tuberculosis peptides.Immunology. 2011 Apr;132(4):482-91. doi: 10.1111/j.1365-2567.2010.03383.x. Epub 2011 Feb 7. Immunology. 2011. PMID: 21294723 Free PMC article.
-
TGFβ-derived immune modulatory vaccine: targeting the immunosuppressive and fibrotic tumor microenvironment in a murine model of pancreatic cancer.J Immunother Cancer. 2022 Dec;10(12):e005491. doi: 10.1136/jitc-2022-005491. J Immunother Cancer. 2022. PMID: 36600556 Free PMC article.
-
Arginase-1-based vaccination against the tumor microenvironment: the identification of an optimal T-cell epitope.Cancer Immunol Immunother. 2019 Nov;68(11):1901-1907. doi: 10.1007/s00262-019-02425-6. Epub 2019 Nov 6. Cancer Immunol Immunother. 2019. PMID: 31690955 Free PMC article. Review.
-
Pro-inflammatory responses after peptide-based cancer immunotherapy.Heliyon. 2024 May 31;10(11):e32249. doi: 10.1016/j.heliyon.2024.e32249. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38912474 Free PMC article. Review.
Cited by
-
Tumor microenvironment antigens.Semin Immunopathol. 2023 Mar;45(2):253-264. doi: 10.1007/s00281-022-00966-0. Epub 2022 Sep 29. Semin Immunopathol. 2023. PMID: 36175673 Free PMC article. Review.
-
Blockade of CCR5+ T Cell Accumulation in the Tumor Microenvironment Optimizes Anti-TGF-β/PD-L1 Bispecific Antibody.Adv Sci (Weinh). 2024 Nov;11(43):e2408598. doi: 10.1002/advs.202408598. Epub 2024 Sep 20. Adv Sci (Weinh). 2024. PMID: 39303165 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
