CycleFlow simultaneously quantifies cell-cycle phase lengths and quiescence in vivo
- PMID: 36313807
- PMCID: PMC9606136
- DOI: 10.1016/j.crmeth.2022.100315
CycleFlow simultaneously quantifies cell-cycle phase lengths and quiescence in vivo
Abstract
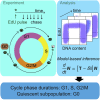
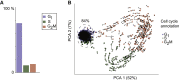
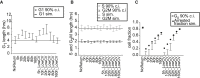
Populations of stem, progenitor, or cancer cells show proliferative heterogeneity in vivo, comprising proliferating and quiescent cells. Consistent quantification of the quiescent subpopulation and progression of the proliferating cells through the individual phases of the cell cycle has not been achieved. Here, we describe CycleFlow, a method that robustly infers this comprehensive information from standard pulse-chase experiments with thymidine analogs. Inference is based on a mathematical model of the cell cycle, with realistic waiting time distributions for the G1, S, and G2/M phases and a long-term quiescent G0 state. We validate CycleFlow with an exponentially growing cancer cell line in vitro. Applying it to T cell progenitors in steady state in vivo, we uncover strong proliferative heterogeneity, with a minority of CD4+CD8+ T cell progenitors cycling very rapidly and then entering quiescence. CycleFlow is suitable as a routine method for quantitative cell-cycle analysis.
Keywords: BrdU labeling; EdU labeling; G0; cell cycle; cell proliferation; cell-cycle arrest; non-Markovian model; quiescence; statistical inference; thymocyte development.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


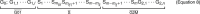
Similar articles
-
Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0 /G1 phase of the cell cycle.Blood. 1997 Dec 1;90(11):4384-93. Blood. 1997. PMID: 9373249
-
SNHG1 opposes quiescence and promotes docetaxel sensitivity in prostate cancer.BMC Cancer. 2023 Jul 18;23(1):672. doi: 10.1186/s12885-023-11006-x. BMC Cancer. 2023. PMID: 37464317 Free PMC article.
-
Cell proliferation kinetics of MCF-7 human mammary carcinoma cells in culture and effects of tamoxifen on exponentially growing and plateau-phase cells.Cancer Res. 1983 Sep;43(9):3998-4006. Cancer Res. 1983. PMID: 6871841
-
Quiescence, an individual journey.Curr Genet. 2019 Jun;65(3):695-699. doi: 10.1007/s00294-018-00928-w. Epub 2019 Jan 16. Curr Genet. 2019. PMID: 30649583 Review.
-
A common strategy for initiating the transition from proliferation to quiescence.Curr Genet. 2017 May;63(2):179-186. doi: 10.1007/s00294-016-0640-0. Epub 2016 Aug 20. Curr Genet. 2017. PMID: 27544284 Free PMC article. Review.
Cited by
-
Harnessing cancer stem cell-derived exosomes to improve cancer therapy.J Exp Clin Cancer Res. 2023 May 23;42(1):131. doi: 10.1186/s13046-023-02717-x. J Exp Clin Cancer Res. 2023. PMID: 37217932 Free PMC article. Review.
-
Interferon regulates neural stem cell function at all ages by orchestrating mTOR and cell cycle.EMBO Mol Med. 2023 Apr 11;15(4):e16434. doi: 10.15252/emmm.202216434. Epub 2023 Jan 13. EMBO Mol Med. 2023. PMID: 36636818 Free PMC article.
References
-
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

