Mirror proteases of Ac-Trypsin and Ac-LysargiNase precisely improve novel event identifications in Mycolicibacterium smegmatis MC2 155 by proteogenomic analysis
- PMID: 36312923
- PMCID: PMC9597629
- DOI: 10.3389/fmicb.2022.1015140
Mirror proteases of Ac-Trypsin and Ac-LysargiNase precisely improve novel event identifications in Mycolicibacterium smegmatis MC2 155 by proteogenomic analysis
Abstract
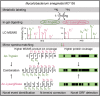
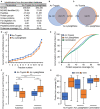
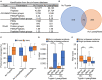
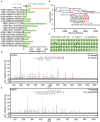
Accurate identification of novel peptides remains challenging because of the lack of evaluation criteria in large-scale proteogenomic studies. Mirror proteases of trypsin and lysargiNase can generate complementary b/y ion series, providing the opportunity to efficiently assess authentic novel peptides in experiments other than filter potential targets by different false discovery rates (FDRs) ranking. In this study, a pair of in-house developed acetylated mirror proteases, Ac-Trypsin and Ac-LysargiNase, were used in Mycolicibacterium smegmatis MC2 155 for proteogenomic analysis. The mirror proteases accurately identified 368 novel peptides, exhibiting 75-80% b and y ion coverages against 65-68% y or b ion coverages of Ac-Trypsin (38.9% b and 68.3% y) or Ac-LysargiNase (65.5% b and 39.6% y) as annotated peptides from M. smegmatis MC2 155. The complementary b and y ion series largely increased the reliability of overlapped sequences derived from novel peptides. Among these novel peptides, 311 peptides were annotated in other public M. smegmatis strains, and 57 novel peptides with more continuous b and y pairs were obtained for further analysis after spectral quality assessment. This enabled mirror proteases to successfully correct six annotated proteins' N-termini and detect 17 new coding open reading frames (ORFs). We believe that mirror proteases will be an effective strategy for novel peptide detection in both prokaryotic and eukaryotic proteogenomics.
Keywords: Ac-LysargiNase; Ac-Trypsin; Mycolicibacterium smegmatis; mirror; proteogenomics.
Copyright © 2022 Jiang, Shi, Li, Zhang, Chang, Wang, Wu, Yu, Dai, Zhang, Lyu, Xu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Ac-LysargiNase efficiently helps genome reannotation of Mycolicibacterium smegmatis MC2 155.J Proteomics. 2022 Jul 30;264:104622. doi: 10.1016/j.jprot.2022.104622. Epub 2022 May 19. J Proteomics. 2022. PMID: 35598869
-
Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics.Mol Cell Proteomics. 2019 Apr;18(4):773-785. doi: 10.1074/mcp.TIR118.000918. Epub 2019 Jan 8. Mol Cell Proteomics. 2019. PMID: 30622160 Free PMC article.
-
Ac-LysargiNase Complements Trypsin for the Identification of Ubiquitinated Sites.Anal Chem. 2019 Dec 17;91(24):15890-15898. doi: 10.1021/acs.analchem.9b04340. Epub 2019 Nov 27. Anal Chem. 2019. PMID: 31774262
-
[Development of LysargiNase, a mirror trypsin and its application in proteomics].Sheng Wu Gong Cheng Xue Bao. 2019 May 25;35(5):741-748. doi: 10.13345/j.cjb.180371. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31222992 Review. Chinese.
-
False discovery rate: the Achilles' heel of proteogenomics.Brief Bioinform. 2022 Sep 20;23(5):bbac163. doi: 10.1093/bib/bbac163. Brief Bioinform. 2022. PMID: 35534181 Review.
Cited by
-
Integration of cloud-based molecular networking and docking for enhanced umami peptide screening from Pixian douban.Food Chem X. 2023 Dec 27;21:101098. doi: 10.1016/j.fochx.2023.101098. eCollection 2024 Mar 30. Food Chem X. 2023. PMID: 38229673 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

