Revisiting influenza A virus life cycle from a perspective of genome balance
- PMID: 36309307
- PMCID: PMC10006207
- DOI: 10.1016/j.virs.2022.10.005
Revisiting influenza A virus life cycle from a perspective of genome balance
Abstract
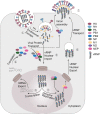
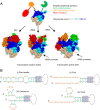
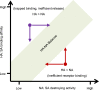
Influenza A virus (IAV) genome comprises eight negative-sense RNA segments, of which the replication is well orchestrated and the delicate balance of multiple segments are dynamically regulated throughout IAV life cycle. However, previous studies seldom discuss these balances except for functional hemagglutinin-neuraminidase balance that is pivotal for both virus entry and release. Therefore, we attempt to revisit IAV life cycle by highlighting the critical role of "genome balance". Moreover, we raise a "balance regression" model of IAV evolution that the virus evolves to rebalance its genome after reassortment or interspecies transmission, and direct a "balance compensation" strategy to rectify the "genome imbalance" as a result of artificial modifications during creation of recombinant IAVs. This review not only improves our understanding of IAV life cycle, but also facilitates both basic and applied research of IAV in future.
Keywords: Balance compensation; Balance regression; Genome balance; Influenza A virus (IAV); Segmented genome.
Copyright © 2022 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Expanding the tolerance of segmented Influenza A Virus genome using a balance compensation strategy.PLoS Pathog. 2022 Aug 4;18(8):e1010756. doi: 10.1371/journal.ppat.1010756. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 35926068 Free PMC article.
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates.Virus Genes. 2022 Aug;58(4):255-269. doi: 10.1007/s11262-022-01904-w. Epub 2022 Apr 26. Virus Genes. 2022. PMID: 35471490 Review.
-
Infection of Mouse Macrophages by Seasonal Influenza Viruses Can Be Restricted at the Level of Virus Entry and at a Late Stage in the Virus Life Cycle.J Virol. 2015 Dec;89(24):12319-29. doi: 10.1128/JVI.01455-15. Epub 2015 Sep 30. J Virol. 2015. PMID: 26423941 Free PMC article.
-
Subtype Diversity of Influenza A Virus in North American Waterfowl: a Multidecade Study.J Virol. 2020 May 18;94(11):e02022-19. doi: 10.1128/JVI.02022-19. Print 2020 May 18. J Virol. 2020. PMID: 32188732 Free PMC article.
-
Roles of Glycans and Non-glycans on the Epithelium and in the Immune System in H1-H18 Influenza A Virus Infections.Methods Mol Biol. 2022;2556:205-242. doi: 10.1007/978-1-0716-2635-1_16. Methods Mol Biol. 2022. PMID: 36175637 Review.
Cited by
-
Carambolaside W Inhibited H1N1 Influenza Virus-Induced Oxidative Stress through STAT-3/BCL-XL Signaling Pathway.Viruses. 2023 Aug 31;15(9):1858. doi: 10.3390/v15091858. Viruses. 2023. PMID: 37766266 Free PMC article.
-
Recent advances of phenotypic screening strategies in the application of anti-influenza virus drug discovery.RSC Med Chem. 2023 Nov 9;15(1):70-80. doi: 10.1039/d3md00513e. eCollection 2024 Jan 25. RSC Med Chem. 2023. PMID: 38283223 Free PMC article. Review.
-
Allopregnanolone targets nucleoprotein as a novel influenza virus inhibitor.Virol Sin. 2023 Dec;38(6):931-939. doi: 10.1016/j.virs.2023.09.003. Epub 2023 Sep 22. Virol Sin. 2023. PMID: 37741571 Free PMC article.
-
Mcl-1 Protein and Viral Infections: A Narrative Review.Int J Mol Sci. 2024 Jan 17;25(2):1138. doi: 10.3390/ijms25021138. Int J Mol Sci. 2024. PMID: 38256213 Free PMC article. Review.
-
The role of ferroptosis in virus infections.Front Microbiol. 2023 Nov 23;14:1279655. doi: 10.3389/fmicb.2023.1279655. eCollection 2023. Front Microbiol. 2023. PMID: 38075884 Free PMC article. Review.
References
-
- Arai Y., Elgendy E.M., Daidoji T., Ibrahim M.S., Ono T., Sriwilaijaroen N., Suzuki Y., Nakaya T., Matsumoto K., Watanabe Y. H9N2 influenza virus infections in human cells require a balance between neuraminidase sialidase activity and hemagglutinin receptor affinity. J. Virol. 2020;94:e01210–e01220. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

