Circr, a Computational Tool to Identify miRNA:circRNA Associations
- PMID: 36304313
- PMCID: PMC9580875
- DOI: 10.3389/fbinf.2022.852834
Circr, a Computational Tool to Identify miRNA:circRNA Associations
Abstract
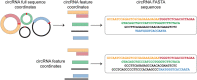
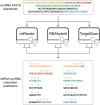
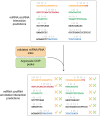
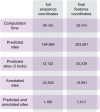
Circular RNAs (circRNAs) are known to act as important regulators of the microRNA (miRNA) activity. Yet, computational resources to identify miRNA:circRNA interactions are mostly limited to already annotated circRNAs or affected by high rates of false positive predictions. To overcome these limitations, we developed Circr, a computational tool for the prediction of associations between circRNAs and miRNAs. Circr combines three publicly available algorithms for de novo prediction of miRNA binding sites on target sequences (miRanda, RNAhybrid, and TargetScan) and annotates each identified miRNA:target pairs with experimentally validated miRNA:RNA interactions and binding sites for Argonaute proteins derived from either ChIPseq or CLIPseq data. The combination of multiple tools for the identification of a single miRNA recognition site with experimental data allows to efficiently prioritize candidate miRNA:circRNA interactions for functional studies in different organisms. Circr can use its internal annotation database or custom annotation tables to enhance the identification of novel and not previously annotated miRNA:circRNA sites in virtually any species. Circr is written in Python 3.6 and is released under the GNU GPL3.0 License at https://github.com/bicciatolab/Circr.
Keywords: circRNA; interactions prediction; miRNA; miRNA binding; prediction tools.
Copyright © 2022 Dori, Caroli and Forcato.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
CircMiMi: a stand-alone software for constructing circular RNA-microRNA-mRNA interactions across species.BMC Bioinformatics. 2022 May 6;23(1):164. doi: 10.1186/s12859-022-04692-0. BMC Bioinformatics. 2022. PMID: 35524165 Free PMC article.
-
Circular RNA expression profile in transgenic diabetic mouse kidneys.Cell Mol Biol Lett. 2021 Jun 7;26(1):25. doi: 10.1186/s11658-021-00270-z. Cell Mol Biol Lett. 2021. PMID: 34098865 Free PMC article.
-
Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations.Genes (Basel). 2019 Aug 24;10(9):642. doi: 10.3390/genes10090642. Genes (Basel). 2019. PMID: 31450634 Free PMC article. Review.
-
CRAFT: a bioinformatics software for custom prediction of circular RNA functions.Brief Bioinform. 2022 Mar 10;23(2):bbab601. doi: 10.1093/bib/bbab601. Brief Bioinform. 2022. PMID: 35106564 Free PMC article.
-
Advances in Circular RNA and Its Applications.Int J Med Sci. 2022 May 27;19(6):975-985. doi: 10.7150/ijms.71840. eCollection 2022. Int J Med Sci. 2022. PMID: 35813288 Free PMC article. Review.
Cited by
-
Adipose Tissue Exosome circ_sxc Mediates the Modulatory of Adiposomes on Brain Aging by Inhibiting Brain dme-miR-87-3p.Mol Neurobiol. 2024 Jan;61(1):224-238. doi: 10.1007/s12035-023-03516-3. Epub 2023 Aug 19. Mol Neurobiol. 2024. PMID: 37597108
-
Investigation of the Circular Transcriptome in Alzheimer's Disease Brain.J Mol Neurosci. 2024 Jul 9;74(3):64. doi: 10.1007/s12031-024-02236-0. J Mol Neurosci. 2024. PMID: 38981928 Free PMC article.
-
CircNetVis: an interactive web application for visualizing interaction networks of circular RNAs.BMC Bioinformatics. 2024 Jan 17;25(1):31. doi: 10.1186/s12859-024-05646-4. BMC Bioinformatics. 2024. PMID: 38233808 Free PMC article.
References
LinkOut - more resources
Full Text Sources

