In Silico Methods for Identification of Potential Active Sites of Therapeutic Targets
- PMID: 36296697
- PMCID: PMC9609013
- DOI: 10.3390/molecules27207103
In Silico Methods for Identification of Potential Active Sites of Therapeutic Targets
Abstract
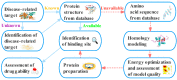
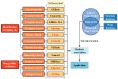
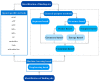
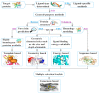
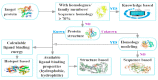
Target identification is an important step in drug discovery, and computer-aided drug target identification methods are attracting more attention compared with traditional drug target identification methods, which are time-consuming and costly. Computer-aided drug target identification methods can greatly reduce the searching scope of experimental targets and associated costs by identifying the diseases-related targets and their binding sites and evaluating the druggability of the predicted active sites for clinical trials. In this review, we introduce the principles of computer-based active site identification methods, including the identification of binding sites and assessment of druggability. We provide some guidelines for selecting methods for the identification of binding sites and assessment of druggability. In addition, we list the databases and tools commonly used with these methods, present examples of individual and combined applications, and compare the methods and tools. Finally, we discuss the challenges and limitations of binding site identification and druggability assessment at the current stage and provide some recommendations and future perspectives.
Keywords: binding site; drug discovery; druggability; target identification; therapeutic target.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Druggability and drug-likeness concepts in drug design: are biomodelling and predictive tools having their say?J Mol Model. 2020 May 8;26(6):120. doi: 10.1007/s00894-020-04385-6. J Mol Model. 2020. PMID: 32382800 Review.
-
Cryptic binding sites on proteins: definition, detection, and druggability.Curr Opin Chem Biol. 2018 Jun;44:1-8. doi: 10.1016/j.cbpa.2018.05.003. Epub 2018 May 23. Curr Opin Chem Biol. 2018. PMID: 29800865 Free PMC article. Review.
-
In Silico Target Druggability Assessment: From Structural to Systemic Approaches.Methods Mol Biol. 2019;1953:63-88. doi: 10.1007/978-1-4939-9145-7_5. Methods Mol Biol. 2019. PMID: 30912016
-
In silico Druggability Assessment of the NUDIX Hydrolase Protein Family as a Workflow for Target Prioritization.Front Chem. 2020 May 29;8:443. doi: 10.3389/fchem.2020.00443. eCollection 2020. Front Chem. 2020. PMID: 32548091 Free PMC article.
-
Binding site druggability assessment in fragment-based drug design.Methods Mol Biol. 2015;1289:13-21. doi: 10.1007/978-1-4939-2486-8_2. Methods Mol Biol. 2015. PMID: 25709029
Cited by
-
In silico identification of putative druggable pockets in PRL3, a significant oncology target.Biochem Biophys Rep. 2024 Jul 1;39:101767. doi: 10.1016/j.bbrep.2024.101767. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39050014 Free PMC article.
-
Integrated Molecular Modeling and Machine Learning for Drug Design.J Chem Theory Comput. 2023 Nov 14;19(21):7478-7495. doi: 10.1021/acs.jctc.3c00814. Epub 2023 Oct 26. J Chem Theory Comput. 2023. PMID: 37883810 Free PMC article. Review.
-
Drug development advances in human genetics-based targets.MedComm (2020). 2024 Feb 9;5(2):e481. doi: 10.1002/mco2.481. eCollection 2024 Feb. MedComm (2020). 2024. PMID: 38344397 Free PMC article. Review.
-
Antimicrobial activity of compounds identified by artificial intelligence discovery engine targeting enzymes involved in Neisseria gonorrhoeae peptidoglycan metabolism.Biol Res. 2024 Sep 5;57(1):62. doi: 10.1186/s40659-024-00543-9. Biol Res. 2024. PMID: 39238057 Free PMC article.
-
Dynamic regulation and key roles of ribonucleic acid methylation.Front Cell Neurosci. 2022 Dec 19;16:1058083. doi: 10.3389/fncel.2022.1058083. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36601431 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

