Estimation of Metabolic Effects upon Cadmium Exposure during Pregnancy Using Tensor Decomposition
- PMID: 36292583
- PMCID: PMC9602417
- DOI: 10.3390/genes13101698
Estimation of Metabolic Effects upon Cadmium Exposure during Pregnancy Using Tensor Decomposition
Abstract
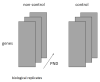
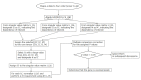
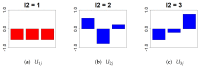
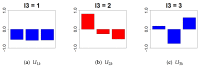
A simple tensor decomposition model was applied to the liver transcriptome analysis data to elucidate the cause of cadmium-induced gene overexpression. In addition, we estimated the mechanism by which prenatal Cd exposure disrupts insulin metabolism in offspring. Numerous studies have reported on the toxicity of Cd. A liver transcriptome analysis revealed that Cd toxicity induces intracellular oxidative stress and mitochondrial dysfunction via changes in gene expression, which in turn induces endoplasmic reticulum-associated degradation via abnormal protein folding. However, the specific mechanisms underlying these effects remain unknown. In this study, we found that Cd-induced endoplasmic reticulum stress may promote increased expression of tumor necrosis factor-α (TNF-α). Based on the high expression of genes involved in the production of sphingolipids, it was also found that the accumulation of ceramide may induce intracellular oxidative stress through the overproduction of reactive oxygen species. In addition, the high expression of a set of genes involved in the electron transfer system may contribute to oxidative stress. These findings allowed us to identify the mechanisms by which intracellular oxidative stress leads to the phosphorylation of insulin receptor substrate 1, which plays a significant role in the insulin signaling pathway.
Keywords: cadmium exposure; insulin metabolism disruption; tensor decomposition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Cadmium exposure suppresses insulin secretion through mtROS-mediated mitochondrial dysfunction and inflammatory response in pancreatic beta cells.J Trace Elem Med Biol. 2022 May;71:126952. doi: 10.1016/j.jtemb.2022.126952. Epub 2022 Feb 16. J Trace Elem Med Biol. 2022. PMID: 35183883
-
Sphingolipids as a Culprit of Mitochondrial Dysfunction in Insulin Resistance and Type 2 Diabetes.Front Endocrinol (Lausanne). 2021 Mar 18;12:635175. doi: 10.3389/fendo.2021.635175. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33815291 Free PMC article. Review.
-
Mitochondrial redox-driven mitofusin 2 S-glutathionylation promotes neuronal necroptosis via disrupting ER-mitochondria crosstalk in cadmium-induced neurotoxicity.Chemosphere. 2021 Jan;262:127878. doi: 10.1016/j.chemosphere.2020.127878. Epub 2020 Aug 6. Chemosphere. 2021. PMID: 33182097
-
Histological changes, lipid metabolism, and oxidative and endoplasmic reticulum stress in the liver of laying hens exposed to cadmium concentrations.Poult Sci. 2020 Jun;99(6):3215-3228. doi: 10.1016/j.psj.2019.12.073. Epub 2020 Mar 5. Poult Sci. 2020. PMID: 32475458 Free PMC article.
-
Mitochondria, reactive oxygen species and cadmium toxicity in the kidney.Toxicol Lett. 2010 Sep 15;198(1):49-55. doi: 10.1016/j.toxlet.2010.04.013. Epub 2010 Apr 22. Toxicol Lett. 2010. PMID: 20417263 Review.
References
-
- Takashige K., Hiroyuki S., Rie F., Yoshito K., Masahisa I., Kojun S., Shinya S., Masao S. Cadmium reduces adipocyte size and expression levels of adiponectin and Peg1/Mest in adipose tissue. Toxicology. 2010;267:20–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

