An integrated multi-omics analysis of topoisomerase family in pan-cancer: Friend or foe?
- PMID: 36288358
- PMCID: PMC9604985
- DOI: 10.1371/journal.pone.0274546
An integrated multi-omics analysis of topoisomerase family in pan-cancer: Friend or foe?
Abstract
Background: Topoisomerases are nuclear enzymes that get to the bottom of topological troubles related with DNA all through a range of genetic procedures. More and more studies have shown that topoisomerase-mediated DNA cleavage plays crucial roles in tumor cell death and carcinogenesis. There is however still a lack of comprehensive multi-omics studies related to topoisomerase family genes from a pan-cancer perspective.
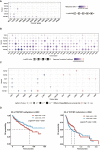
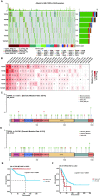
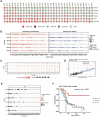
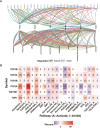
Methods: In this study, a multiomics pan-cancer analysis of topoisomerase family genes was conducted by integrating over 10,000 multi-dimensional cancer genomic data across 33 cancer types from The Cancer Genome Atlas (TCGA), 481 small molecule drug response data from cancer therapeutics response portal (CTRP) as well as normal tissue data from Genotype-Tissue Expression (GTEx). Finally, overall activity-level analyses of topoisomerase in pan-cancers were performed by gene set variation analysis (GSVA), together with differential expression, clinical relevancy, immune cell infiltration and regulation of cancer-related pathways.
Results: Dysregulated gene expression of topoisomerase family were related to genomic changes and abnormal epigenetic modifications. The expression levels of topoisomerase family genes could significantly impact cancer progression, intratumoral heterogeneity, alterations in the immunological condition and regulation of the cancer marker-related pathways, which in turn caused the differences in potential drugs sensitivity and the distinct prognosis of patients.
Conclusion: It was anticipated that topoisomerase family genes would become novel prognostic biomarkers for cancer patients and provide new insights for the diagnosis and treatment of tumors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Multi-omics analysis reveals the panoramic picture of necroptosis-related regulators in pan-cancer.Aging (Albany NY). 2022 Jun 21;14(12):5034-5058. doi: 10.18632/aging.204124. Epub 2022 Jun 21. Aging (Albany NY). 2022. PMID: 35748782 Free PMC article.
-
Comprehensive pan-cancer analysis of inflammatory age-clock-related genes as prognostic and immunity markers based on multi-omics data.Sci Rep. 2024 May 7;14(1):10468. doi: 10.1038/s41598-024-61381-z. Sci Rep. 2024. PMID: 38714870 Free PMC article.
-
Pan-Cancer Analysis Reveals Genomic and Clinical Characteristics of TRPV Channel-Related Genes.Front Oncol. 2022 Jan 31;12:813100. doi: 10.3389/fonc.2022.813100. eCollection 2022. Front Oncol. 2022. PMID: 35174089 Free PMC article.
-
A comprehensive overview of oncogenic pathways in human cancer.Brief Bioinform. 2020 May 21;21(3):957-969. doi: 10.1093/bib/bbz046. Brief Bioinform. 2020. PMID: 31155677 Review.
-
Onco-Multi-OMICS Approach: A New Frontier in Cancer Research.Biomed Res Int. 2018 Oct 3;2018:9836256. doi: 10.1155/2018/9836256. eCollection 2018. Biomed Res Int. 2018. PMID: 30402498 Free PMC article. Review.
Cited by
-
Exploration of potential molecular mechanisms and genotoxicity of anti-cancer drugs using next generation knowledge discovery methods.Pak J Med Sci. 2023 Jul-Aug;39(4):988-993. doi: 10.12669/pjms.39.4.7427. Pak J Med Sci. 2023. PMID: 37492288 Free PMC article.
-
Understanding Cancer's Defense against Topoisomerase-Active Drugs: A Comprehensive Review.Cancers (Basel). 2024 Feb 6;16(4):680. doi: 10.3390/cancers16040680. Cancers (Basel). 2024. PMID: 38398072 Free PMC article. Review.
-
Evaluation of the Anticancer and Biological Activities of Istaroxime via Ex Vivo Analyses, Molecular Docking and Conceptual Density Functional Theory Computations.Molecules. 2023 Nov 7;28(22):7458. doi: 10.3390/molecules28227458. Molecules. 2023. PMID: 38005181 Free PMC article.
References
-
- Song J, Song F, Liu K, Zhang W, Luo R, Tang Y, et al.. Multi-omics analysis reveals epithelial-mesenchymal transition-related gene FOXM1 as a novel prognostic biomarker in clear cell renal carcinoma. Aging (Albany NY). 2019;11(22):10316–37. Epub 2019/11/20. doi: 10.18632/aging.102459 ; PubMed Central PMCID: PMC6914426. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

