In Vitro Demonstration of Human Lipoyl Synthase Catalytic Activity in the Presence of NFU1
- PMID: 36281303
- PMCID: PMC9585516
- DOI: 10.1021/acsbiomedchemau.2c00020
In Vitro Demonstration of Human Lipoyl Synthase Catalytic Activity in the Presence of NFU1
Abstract
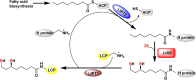
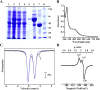
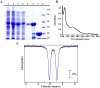
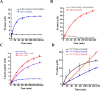
Lipoyl synthase (LS) catalyzes the last step in the biosynthesis of the lipoyl cofactor, which is the attachment of sulfur atoms at C6 and C8 of an n-octanoyllysyl side chain of a lipoyl carrier protein (LCP). The protein is a member of the radical S-adenosylmethionine (SAM) superfamily of enzymes, which use SAM as a precursor to a 5'-deoxyadenosyl 5'-radical (5'-dA·). The role of the 5'-dA· in the LS reaction is to abstract hydrogen atoms from C6 and C8 of the octanoyl moiety of the substrate to initiate subsequent sulfur attachment. All radical SAM enzymes have at least one [4Fe-4S] cluster that is used in the reductive cleavage of SAM to generate the 5'-dA·; however, LSs contain an additional auxiliary [4Fe-4S] cluster from which sulfur atoms are extracted during turnover, leading to degradation of the cluster. Therefore, these enzymes catalyze only 1 turnover in the absence of a system that restores the auxiliary cluster. In Escherichia coli, the auxiliary cluster of LS can be regenerated by the iron-sulfur (Fe-S) cluster carrier protein NfuA as fast as catalysis takes place, and less efficiently by IscU. NFU1 is the human ortholog of E. coli NfuA and has been shown to interact directly with human LS (i.e., LIAS) in yeast two-hybrid analyses. Herein, we show that NFU1 and LIAS form a tight complex in vitro and that NFU1 can efficiently restore the auxiliary cluster of LIAS during turnover. We also show that BOLA3, previously identified as being critical in the biosynthesis of the lipoyl cofactor in humans and Saccharomyces cerevisiae, has no direct effect on Fe-S cluster transfer from NFU1 or GLRX5 to LIAS. Further, we show that ISCA1 and ISCA2 can enhance LIAS turnover, but only slightly.
© 2022 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
The A-type domain in Escherichia coli NfuA is required for regenerating the auxiliary [4Fe-4S] cluster in Escherichia coli lipoyl synthase.J Biol Chem. 2019 Feb 1;294(5):1609-1617. doi: 10.1074/jbc.RA118.006171. Epub 2018 Dec 11. J Biol Chem. 2019. PMID: 30538130 Free PMC article.
-
Biochemical Approaches to Probe the Role of the Auxiliary Iron-Sulfur Cluster of Lipoyl Synthase from Mycobacterium Tuberculosis.Methods Mol Biol. 2021;2353:307-332. doi: 10.1007/978-1-0716-1605-5_16. Methods Mol Biol. 2021. PMID: 34292556
-
Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase.Biochemistry. 2014 Jul 22;53(28):4557-72. doi: 10.1021/bi500432r. Epub 2014 Jul 10. Biochemistry. 2014. PMID: 24901788 Free PMC article.
-
Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review.
-
Auxiliary iron-sulfur cofactors in radical SAM enzymes.Biochim Biophys Acta. 2015 Jun;1853(6):1316-34. doi: 10.1016/j.bbamcr.2015.01.002. Epub 2015 Jan 15. Biochim Biophys Acta. 2015. PMID: 25597998 Review.
Cited by
-
Twenty Years of Radical SAM! The Genesis of the Superfamily.ACS Bio Med Chem Au. 2022 Dec 5;2(6):538-547. doi: 10.1021/acsbiomedchemau.2c00078. eCollection 2022 Dec 21. ACS Bio Med Chem Au. 2022. PMID: 37101427 Free PMC article. No abstract available.
-
Lipoic acid attachment to proteins: stimulating new developments.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0000524. doi: 10.1128/mmbr.00005-24. Epub 2024 Apr 16. Microbiol Mol Biol Rev. 2024. PMID: 38624243 Review.
-
FDX1 regulates cellular protein lipoylation through direct binding to LIAS.bioRxiv [Preprint]. 2023 Feb 4:2023.02.03.526472. doi: 10.1101/2023.02.03.526472. bioRxiv. 2023. Update in: J Biol Chem. 2023 Sep;299(9):105046. doi: 10.1016/j.jbc.2023.105046 PMID: 36778498 Free PMC article. Updated. Preprint.
References
-
- Patterson E. L.; et al. Crystallization of a Derivative of Protogen-B. J. Am. Chem. Soc. 1951, 73 (12), 5919–5920. 10.1021/ja01156a566. - DOI
-
- Reed L.Advances in Enzymology and Related Areas of Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, 2006; pp 319–347.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
