Use of compressed sensing to expedite high-throughput diagnostic testing for COVID-19 and beyond
- PMID: 36279287
- PMCID: PMC9632879
- DOI: 10.1371/journal.pcbi.1010629
Use of compressed sensing to expedite high-throughput diagnostic testing for COVID-19 and beyond
Abstract
The rapid spread of SARS-CoV-2 has placed a significant burden on public health systems to provide swift and accurate diagnostic testing highlighting the critical need for innovative testing approaches for future pandemics. In this study, we present a novel sample pooling procedure based on compressed sensing theory to accurately identify virally infected patients at high prevalence rates utilizing an innovative viral RNA extraction process to minimize sample dilution. At prevalence rates ranging from 0-14.3%, the number of tests required to identify the infection status of all patients was reduced by 69.26% as compared to conventional testing in primary human SARS-CoV-2 nasopharyngeal swabs and a coronavirus model system. Our method provided quantification of individual sample viral load within a pool as well as a binary positive-negative result. Additionally, our modified pooling and RNA extraction process minimized sample dilution which remained constant as pool sizes increased. Compressed sensing can be adapted to a wide variety of diagnostic testing applications to increase throughput for routine laboratory testing as well as a means to increase testing capacity to combat future pandemics.
Conflict of interest statement
The authors are coinventors of a pending patent covering the use of compressed sensing in diagnostic testing applications.
Figures
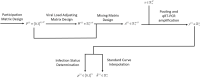

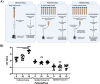
Update of
-
Use of compressed sensing to expedite high-throughput diagnostic testing for COVID-19 and beyond.medRxiv [Preprint]. 2021 Aug 10:2021.08.09.21261669. doi: 10.1101/2021.08.09.21261669. medRxiv. 2021. Update in: PLoS Comput Biol. 2022 Oct 24;18(10):e1010629. doi: 10.1371/journal.pcbi.1010629. PMID: 34401889 Free PMC article. Updated. Preprint.
Similar articles
-
Use of compressed sensing to expedite high-throughput diagnostic testing for COVID-19 and beyond.medRxiv [Preprint]. 2021 Aug 10:2021.08.09.21261669. doi: 10.1101/2021.08.09.21261669. medRxiv. 2021. Update in: PLoS Comput Biol. 2022 Oct 24;18(10):e1010629. doi: 10.1371/journal.pcbi.1010629. PMID: 34401889 Free PMC article. Updated. Preprint.
-
High-Throughput COVID-19 Testing of Naso-Oropharyngeal Swabs Using a Sensitive Extraction-Free Sample Preparation Method.Microbiol Spectr. 2022 Aug 31;10(4):e0135822. doi: 10.1128/spectrum.01358-22. Epub 2022 Aug 11. Microbiol Spectr. 2022. PMID: 35950846 Free PMC article.
-
Critical Aspects Concerning the Development of a Pooling Approach for SARS-CoV-2 Diagnosis Using Large-Scale PCR Testing.Viruses. 2021 May 13;13(5):902. doi: 10.3390/v13050902. Viruses. 2021. PMID: 34067983 Free PMC article.
-
Current Status of Diagnostic Testing for SARS-CoV-2 Infection and Future Developments: A Review.Med Sci Monit. 2020 Dec 17;26:e928552. doi: 10.12659/MSM.928552. Med Sci Monit. 2020. PMID: 33332288 Free PMC article. Review.
-
[SARS-CoV-2 and Microbiological Diagnostic Dynamics in COVID-19 Pandemic].Mikrobiyol Bul. 2020 Jul;54(3):497-509. doi: 10.5578/mb.69839. Mikrobiyol Bul. 2020. PMID: 32755524 Review. Turkish.
References
-
- COVID-19 Response. COVID-19 Case Surveillance Public Data Access, Summary, and Limitations (version date: May 5, 2022). Centers for Disease Control and Prevention.
-
- Daily State-by-State Testing Trends. Johns Hopkins Coronavirus Resource Center.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

