CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses
- PMID: 36279285
- PMCID: PMC9632919
- DOI: 10.1371/journal.ppat.1010479
CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses
Abstract
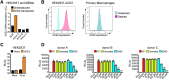
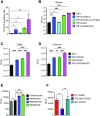
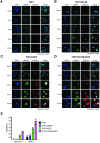
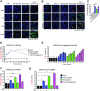
Exacerbated and persistent innate immune response marked by pro-inflammatory cytokine expression is thought to be a major driver of chronic COVID-19 pathology. Although macrophages are not the primary target cells of SARS-CoV-2 infection in humans, viral RNA and antigens in activated monocytes and macrophages have been detected in post-mortem samples, and dysfunctional monocytes and macrophages have been hypothesized to contribute to a protracted hyper-inflammatory state in COVID-19 patients. In this study, we demonstrate that CD169, a myeloid cell specific I-type lectin, facilitated ACE2-independent SARS-CoV-2 fusion and entry in macrophages. CD169-mediated SARS-CoV-2 entry in macrophages resulted in expression of viral genomic and subgenomic RNAs with minimal viral protein expression and no infectious viral particle release, suggesting a post-entry restriction of the SARS-CoV-2 replication cycle. Intriguingly this post-entry replication block was alleviated by exogenous ACE2 expression in macrophages. Restricted expression of viral genomic and subgenomic RNA in CD169+ macrophages elicited a pro-inflammatory cytokine expression (TNFα, IL-6 and IL-1β) in a RIG-I, MDA-5 and MAVS-dependent manner, which was suppressed by remdesivir treatment. These findings suggest that de novo expression of SARS-CoV-2 RNA in macrophages contributes to the pro-inflammatory cytokine signature and that blocking CD169-mediated ACE2 independent infection and subsequent activation of macrophages by viral RNA might alleviate COVID-19-associated hyperinflammatory response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Update of
-
CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses.bioRxiv [Preprint]. 2022 Mar 30:2022.03.29.486190. doi: 10.1101/2022.03.29.486190. bioRxiv. 2022. Update in: PLoS Pathog. 2022 Oct 24;18(10):e1010479. doi: 10.1371/journal.ppat.1010479 PMID: 35378756 Free PMC article. Updated. Preprint.
Similar articles
-
CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses.bioRxiv [Preprint]. 2022 Mar 30:2022.03.29.486190. doi: 10.1101/2022.03.29.486190. bioRxiv. 2022. Update in: PLoS Pathog. 2022 Oct 24;18(10):e1010479. doi: 10.1371/journal.ppat.1010479 PMID: 35378756 Free PMC article. Updated. Preprint.
-
SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages.Front Immunol. 2021 Jun 23;12:683800. doi: 10.3389/fimmu.2021.683800. eCollection 2021. Front Immunol. 2021. PMID: 34248968 Free PMC article.
-
ACE2-independent SARS-CoV-2 virus entry through cell surface GRP78 on monocytes - evidence from a translational clinical and experimental approach.EBioMedicine. 2023 Dec;98:104869. doi: 10.1016/j.ebiom.2023.104869. Epub 2023 Nov 13. EBioMedicine. 2023. PMID: 37967509 Free PMC article.
-
Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D.Int J Mol Sci. 2021 May 16;22(10):5251. doi: 10.3390/ijms22105251. Int J Mol Sci. 2021. PMID: 34065735 Free PMC article. Review.
-
Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions.Life Sci. 2020 Sep 15;257:118102. doi: 10.1016/j.lfs.2020.118102. Epub 2020 Jul 18. Life Sci. 2020. PMID: 32687918 Free PMC article. Review.
Cited by
-
Effects of Different Types of Recombinant SARS-CoV-2 Spike Protein on Circulating Monocytes' Structure.Int J Mol Sci. 2023 May 27;24(11):9373. doi: 10.3390/ijms24119373. Int J Mol Sci. 2023. PMID: 37298324 Free PMC article.
-
Exacerbation of Darier's disease with COVID-19.JAAD Case Rep. 2022 Sep;27:64-66. doi: 10.1016/j.jdcr.2022.06.037. Epub 2022 Jul 6. JAAD Case Rep. 2022. PMID: 35818536 Free PMC article. No abstract available.
-
Modulations of Homeostatic ACE2, CD147, GRP78 Pathways Correlate with Vascular and Endothelial Performance Markers during Pulmonary SARS-CoV-2 Infection.Cells. 2024 Feb 29;13(5):432. doi: 10.3390/cells13050432. Cells. 2024. PMID: 38474396 Free PMC article.
-
Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation.Cell Mol Immunol. 2023 Jul;20(7):835-849. doi: 10.1038/s41423-023-01039-4. Epub 2023 May 30. Cell Mol Immunol. 2023. PMID: 37253946 Free PMC article.
-
Dissecting the abilities of murine Siglecs to interact with gangliosides.J Biol Chem. 2024 Jul;300(7):107482. doi: 10.1016/j.jbc.2024.107482. Epub 2024 Jun 17. J Biol Chem. 2024. PMID: 38897567 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

