Novel Deep Eutectic Solvent-Based Protein Extraction Method for Pottery Residues and Archeological Implications
- PMID: 36268809
- PMCID: PMC9639204
- DOI: 10.1021/acs.jproteome.2c00340
Novel Deep Eutectic Solvent-Based Protein Extraction Method for Pottery Residues and Archeological Implications
Abstract
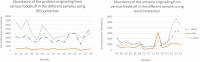
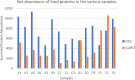
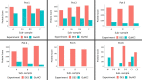
Proteomic analysis of absorbed residues is increasingly used to identify the foodstuffs processed in ancient ceramic vessels, but detailed methodological investigations in this field remain rare. Here, we present three interlinked methodological developments with important consequences in paleoproteomics: the comparative absorption and identification of various food proteins, the application of a deep eutectic solvent (DES) for extracting ceramic-bound proteins, and the role of database choice in taxonomic identification. Our experiments with modern and ethnoarcheological ceramics show that DES is generally more effective at extracting ceramic-bound proteins than guanidine hydrochloride (GuHCl), and cereal proteins are absorbed and subsequently extracted and identifiedat least as readily as meat proteins. We also highlight some of the challenges in cross-species proteomics, whereby species that are less well-represented in databases can be attributed an incorrect species-level taxonomic assignment due to interspecies similarities in protein sequence. This is particularly problematic in potentially mixed samples such as cooking-generated organic residues deposited in pottery. Our work demonstrates possible proteomic separation of fishes and birds, the latter of which have so far eluded detection through lipidomic analyses of organic residue deposits in pottery, which has important implications for tracking the exploitation of avian species in various ancient communities around the globe.
Keywords: Archaeological pottery; Cross-species proteomics; DES; Deep eutectic solvent; Protein extraction; Residue analysis.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


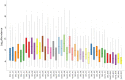

Similar articles
-
Paleoproteomic profiling of organic residues on prehistoric pottery from Malta.Amino Acids. 2021 Feb;53(2):295-312. doi: 10.1007/s00726-021-02946-4. Epub 2021 Feb 13. Amino Acids. 2021. PMID: 33582869 Free PMC article.
-
Accurate compound-specific 14C dating of archaeological pottery vessels.Nature. 2020 Apr;580(7804):506-510. doi: 10.1038/s41586-020-2178-z. Epub 2020 Apr 8. Nature. 2020. PMID: 32322061
-
Deep eutectic solvents as new media for green extraction of food proteins: Opportunity and challenges.Food Res Int. 2022 Nov;161:111842. doi: 10.1016/j.foodres.2022.111842. Epub 2022 Aug 28. Food Res Int. 2022. PMID: 36192972 Review.
-
Different Analytical Procedures for the Study of Organic Residues in Archeological Ceramic Samples with the Use of Gas Chromatography-mass Spectrometry.Crit Rev Anal Chem. 2016;46(1):67-81. doi: 10.1080/10408347.2015.1008130. Crit Rev Anal Chem. 2016. PMID: 25830900 Review.
-
Fine Endmesolithic fish caviar meal discovered by proteomics in foodcrusts from archaeological site Friesack 4 (Brandenburg, Germany).PLoS One. 2018 Nov 28;13(11):e0206483. doi: 10.1371/journal.pone.0206483. eCollection 2018. PLoS One. 2018. PMID: 30485287 Free PMC article.
Cited by
-
The impact of cooking and burial on proteins: a characterisation of experimental foodcrusts and ceramics.R Soc Open Sci. 2024 Sep 18;11(9):240610. doi: 10.1098/rsos.240610. eCollection 2024 Sep. R Soc Open Sci. 2024. PMID: 39416716 Free PMC article.
-
The potential of aptamers for the analysis of ceramic bound proteins found within pottery.Sci Rep. 2024 Aug 27;14(1):19947. doi: 10.1038/s41598-024-70048-8. Sci Rep. 2024. PMID: 39198509 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

