Sorting nexin-dependent therapeutic targeting of oncogenic epidermal growth factor receptor
- PMID: 36253541
- PMCID: PMC9935382
- DOI: 10.1038/s41417-022-00541-7
Sorting nexin-dependent therapeutic targeting of oncogenic epidermal growth factor receptor
Abstract
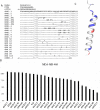
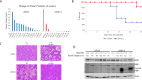
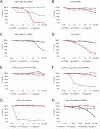
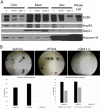
Overexpression and/or overactivation of the Epidermal Growth Factor Receptor (EGFR) is oncogenic in several tumor types yet targeting the kinase domain of wildtype EGFR has had limited success. EGFR has numerous kinase-independent roles, one of which is accomplished through the Sorting Nexin-dependent retrotranslocation of EGFR to the nucleus, which is observed in some metastatic cancers and therapeutically resistant disease. Here, we have utilized the BAR domain of Sorting Nexin 1 to create a peptide-based therapeutic (cSNX1.3) that promotes cell death in EGFR-expressing cancer. We evaluated the efficacy of cSNX1.3 in tumor-bearing WAP-TGFα transgenic mice (an EGFR-dependent model of breast cancer), where cSNX1.3 treatment resulted in significant tumor regression without observable toxicity. Evaluation of remaining tumor tissues found evidence of increased PARP cleavage, suggesting apoptotic tumor cell death. To evaluate the mechanism of action for cSNX1.3, we found that cSNX1.3 binds the C-terminus of the EGFR kinase domain at an interface site opposite the ATP binding domain with a Kd of ~4.0 µM. In vitro analysis found that cSNX1.3 inhibits the nuclear localization of EGFR. To determine specificity, we evaluated cancer cell lines expressing wildtype EGFR (MDA-MB-468, BT20 and A549), mutant EGFR (H1975) and non-transformed lines (CHO and MCF10A). Only transformed lines expressing wildtype EGFR responded to cSNX1.3, while mutant EGFR and normal cells responded better to an EGFR kinase inhibitor. Phenotypically, cSNX1.3 inhibits EGF-, NRG-, and HGF-dependent migration, but not HA-dependent migration. Together, these data indicate that targeting retrotranslocation of EGFR may be a potent therapeutic for RTK-active cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Nuclear EGFR in breast cancer suppresses NK cell recruitment and cytotoxicity.Oncogene. 2024 Nov 9. doi: 10.1038/s41388-024-03211-0. Online ahead of print. Oncogene. 2024. PMID: 39521886
-
Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab.BMC Med. 2012 Mar 21;10:28. doi: 10.1186/1741-7015-10-28. BMC Med. 2012. PMID: 22436374 Free PMC article.
-
Stapled EGFR peptide reduces inflammatory breast cancer and inhibits additional HER-driven models of cancer.J Transl Med. 2019 Jun 18;17(1):201. doi: 10.1186/s12967-019-1939-7. J Transl Med. 2019. PMID: 31215437 Free PMC article.
-
ErbB-2 blockade and prenyltransferase inhibition alter epidermal growth factor and epidermal growth factor receptor trafficking and enhance (111)In-DTPA-hEGF Auger electron radiation therapy.J Nucl Med. 2011 May;52(5):776-83. doi: 10.2967/jnumed.110.084392. Epub 2011 Apr 15. J Nucl Med. 2011. PMID: 21498540
-
The ErbB/HER family of protein-tyrosine kinases and cancer.Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov 20. Pharmacol Res. 2014. PMID: 24269963 Review.
Cited by
-
Expression Profiling of EMT Transcriptional Regulators ZEB1 and ZEB2 in Different Histopathological Grades of Oral Squamous Cell Carcinoma Patients.Curr Genomics. 2024 Apr 8;25(2):140-151. doi: 10.2174/0113892029284920240212091903. Curr Genomics. 2024. PMID: 38751602 Free PMC article.
-
Nuclear epidermal growth factor receptor as a therapeutic target.Explor Target Antitumor Ther. 2023;4(4):616-629. doi: 10.37349/etat.2023.00156. Epub 2023 Aug 30. Explor Target Antitumor Ther. 2023. PMID: 37720348 Free PMC article. Review.
-
Nuclear EGFR in breast cancer suppresses NK cell recruitment and cytotoxicity.Oncogene. 2024 Nov 9. doi: 10.1038/s41388-024-03211-0. Online ahead of print. Oncogene. 2024. PMID: 39521886
-
A peptide derived from sorting nexin 1 inhibits HPV16 entry, retrograde trafficking, and L2 membrane spanning.Tumour Virus Res. 2024 Dec;18:200287. doi: 10.1016/j.tvr.2024.200287. Epub 2024 Jun 21. Tumour Virus Res. 2024. PMID: 38909779 Free PMC article.
-
Spatiotemporal regulation of the hepatocyte growth factor receptor MET activity by sorting nexins 1/2 in HCT116 colorectal cancer cells.Biosci Rep. 2024 Jun 26;44(6):BSR20240182. doi: 10.1042/BSR20240182. Biosci Rep. 2024. PMID: 38836326 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

