Liquid-liquid phase separation mediates the formation of herpesvirus assembly compartments
- PMID: 36250941
- PMCID: PMC9579985
- DOI: 10.1083/jcb.202201088
Liquid-liquid phase separation mediates the formation of herpesvirus assembly compartments
Abstract
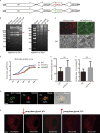
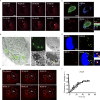
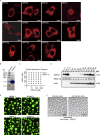
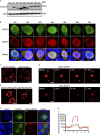
Virus assembly, which takes place during the late stage of viral replication, is essential for virus propagation. However, the underlying mechanisms remain poorly understood, especially for viruses with complicated structures. Here, we use correlative light and electron microscopy to examine the formation of cytoplasmic virion assembly compartments (cVACs) during infection by a γ-herpesvirus. These cVACs are membraneless organelles with liquid-like properties. Formation of cVACs during virus infection is mediated by ORF52, an abundant tegument protein. ORF52 undergoes liquid-liquid phase separation (LLPS), which is promoted by both DNA and RNA. Disrupting ORF52 phase separation blocks cVACs formation and virion production. These results demonstrate that phase separation of ORF52 is critical for cVACs formation. Our work defines herpesvirus cVACs as membraneless compartments that are generated through a process of LLPS mediated by a tegument protein and adds to the cellular processes that are facilitated by phase separation.
© 2022 Zhou et al.
Figures



Comment in
-
Liquid-liquid phase separation drives herpesvirus assembly in the cytoplasm.J Cell Biol. 2023 Jan 2;222(1):e202211015. doi: 10.1083/jcb.202211015. Epub 2022 Dec 21. J Cell Biol. 2023. PMID: 36542408 Free PMC article.
Similar articles
-
Liquid-liquid phase separation drives herpesvirus assembly in the cytoplasm.J Cell Biol. 2023 Jan 2;222(1):e202211015. doi: 10.1083/jcb.202211015. Epub 2022 Dec 21. J Cell Biol. 2023. PMID: 36542408 Free PMC article.
-
Maturation and vesicle-mediated egress of primate gammaherpesvirus rhesus monkey rhadinovirus require inner tegument protein ORF52.J Virol. 2014 Aug;88(16):9111-28. doi: 10.1128/JVI.01502-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899183 Free PMC article.
-
Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm.J Virol. 2007 Sep;81(18):10137-50. doi: 10.1128/JVI.01233-06. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634243 Free PMC article.
-
When liquid-liquid phase separation meets viral infections.Front Immunol. 2022 Aug 9;13:985622. doi: 10.3389/fimmu.2022.985622. eCollection 2022. Front Immunol. 2022. PMID: 36016945 Free PMC article. Review.
-
Herpesvirus assembly: a tale of two membranes.Curr Opin Microbiol. 2006 Aug;9(4):423-9. doi: 10.1016/j.mib.2006.06.013. Epub 2006 Jun 30. Curr Opin Microbiol. 2006. PMID: 16814597 Review.
Cited by
-
Aurora-A condensation mediated by BuGZ aids its mitotic centrosome functions.iScience. 2024 Apr 18;27(5):109785. doi: 10.1016/j.isci.2024.109785. eCollection 2024 May 17. iScience. 2024. PMID: 38746663 Free PMC article.
-
Liquid-liquid phase separation is essential for reovirus viroplasm formation and immune evasion.J Virol. 2024 Sep 17;98(9):e0102824. doi: 10.1128/jvi.01028-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194247 Free PMC article.
-
Cyclic GMP-AMP synthase recognizes the physical features of DNA.Acta Pharmacol Sin. 2025 Feb;46(2):264-270. doi: 10.1038/s41401-024-01369-7. Epub 2024 Aug 7. Acta Pharmacol Sin. 2025. PMID: 39112770 Review.
-
Liquid-liquid phase separation: a new perspective on respiratory diseases.Front Immunol. 2024 Sep 26;15:1444253. doi: 10.3389/fimmu.2024.1444253. eCollection 2024. Front Immunol. 2024. PMID: 39391315 Free PMC article. Review.
-
Advances in the immunoescape mechanisms exploited by alphaherpesviruses.Front Microbiol. 2024 Jun 19;15:1392814. doi: 10.3389/fmicb.2024.1392814. eCollection 2024. Front Microbiol. 2024. PMID: 38962133 Free PMC article. Review.
References
-
- Benach, J., Wang L., Chen Y., Ho C.K., Lee S., Seetharaman J., Xiao R., Acton T.B., Montelione G.T., Deng H., et al. . 2007. Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68. J. Biol. Chem. 282:31534–31541. 10.1074/jbc.M705637200 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

