Interaction of BES1 and LBD37 transcription factors modulates brassinosteroid-regulated root forging response under low nitrogen in arabidopsis
- PMID: 36247555
- PMCID: PMC9555238
- DOI: 10.3389/fpls.2022.998961
Interaction of BES1 and LBD37 transcription factors modulates brassinosteroid-regulated root forging response under low nitrogen in arabidopsis
Abstract
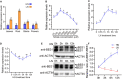
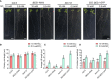
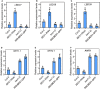
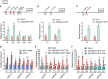
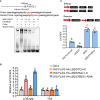
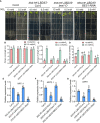
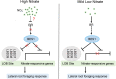
Brassinosteriod (BR) plays important roles in regulation of plant growth, development and environmental responses. BR signaling regulates multiple biological processes through controlling the activity of BES1/BZR1 regulators. Apart from the roles in the promotion of plant growth, BR is also involved in regulation of the root foraging response under low nitrogen, however how BR signaling regulate this process remains unclear. Here we show that BES1 and LBD37 antagonistically regulate root foraging response under low nitrogen conditions. Both the transcriptional level and dephosphorylated level of BES1, is significant induced by low nitrogen, predominantly in root. Phenotypic analysis showed that BES1 gain-of-function mutant or BES1 overexpression transgenic plants exhibits progressive outgrowth of lateral root in response to low nitrogen and BES1 negatively regulates repressors of nitrate signaling pathway and positively regulates several key genes required for NO3 - uptake and signaling. In contrast, BES1 knock-down mutant BES1-RNAi exhibited a dramatical reduction of lateral root elongation in response to low N. Furthermore, we identified a BES1 interacting protein, LBD37, which is a negative repressor of N availability signals. Our results showed that BES1 can inhibit LBD37 transcriptional repression on N-responsive genes. Our results thus demonstrated that BES1-LBD37 module acts critical nodes to integrate BR signaling and nitrogen signaling to modulate the root forging response at LN condition.
Keywords: BES1 transcription factor; LBD37; brassinosteriods; low nitrogen; root forging response.
Copyright © 2022 Chai, Chen, Yue, Li, Zhang, de Dios, Yao and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
BZR1 and BES1 transcription factors mediate brassinosteroid control over root system architecture in response to nitrogen availability.Front Plant Sci. 2024 May 8;15:1387321. doi: 10.3389/fpls.2024.1387321. eCollection 2024. Front Plant Sci. 2024. PMID: 38779077 Free PMC article.
-
Transcriptional network regulation of the brassinosteroid signaling pathway by the BES1-TPL-HDA19 co-repressor complex.Planta. 2019 Oct;250(4):1371-1377. doi: 10.1007/s00425-019-03233-z. Epub 2019 Jul 6. Planta. 2019. PMID: 31280329
-
MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis.Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):20142-7. doi: 10.1073/pnas.1205232109. Epub 2012 Nov 19. Proc Natl Acad Sci U S A. 2012. PMID: 23169658 Free PMC article.
-
The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth.Biochim Biophys Acta Gene Regul Mech. 2018 Jun;1861(6):561-571. doi: 10.1016/j.bbagrm.2018.04.003. Epub 2018 Apr 17. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29673687 Review.
-
Review: Emerging roles of brassinosteroid in nutrient foraging.Plant Sci. 2020 Jul;296:110474. doi: 10.1016/j.plantsci.2020.110474. Epub 2020 Mar 20. Plant Sci. 2020. PMID: 32540004 Review.
Cited by
-
Crosstalk between brassinosteroid signaling and variable nutrient environments.Sci China Life Sci. 2023 Jun;66(6):1231-1244. doi: 10.1007/s11427-022-2319-0. Epub 2023 Mar 9. Sci China Life Sci. 2023. PMID: 36907968 Review.
-
Transcriptome Profiling Reveals the Gene Network Responding to Low Nitrogen Stress in Wheat.Plants (Basel). 2024 Jan 26;13(3):371. doi: 10.3390/plants13030371. Plants (Basel). 2024. PMID: 38337903 Free PMC article.
-
Insights on Phytohormonal Crosstalk in Plant Response to Nitrogen Stress: A Focus on Plant Root Growth and Development.Int J Mol Sci. 2023 Feb 11;24(4):3631. doi: 10.3390/ijms24043631. Int J Mol Sci. 2023. PMID: 36835044 Free PMC article. Review.
-
The heat response regulators HSFA1s promote Arabidopsis thermomorphogenesis via stabilizing PIF4 during the day.Sci Adv. 2023 Nov 3;9(44):eadh1738. doi: 10.1126/sciadv.adh1738. Epub 2023 Nov 3. Sci Adv. 2023. PMID: 37922351 Free PMC article.
-
Multi-Omics Analysis Reveals Synergistic Enhancement of Nitrogen Assimilation Efficiency via Coordinated Regulation of Nitrogen and Carbon Metabolism by Co-Application of Brassinolide and Pyraclostrobin in Arabidopsis thaliana.Int J Mol Sci. 2023 Nov 17;24(22):16435. doi: 10.3390/ijms242216435. Int J Mol Sci. 2023. PMID: 38003624 Free PMC article.
References
-
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., et al. . (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. 111, 2029–2034. doi: 10.1073/pnas.1319953111 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

