miRNA mediated downregulation of cyclase-associated protein 1 (CAP1) is required for myoblast fusion
- PMID: 36246999
- PMCID: PMC9562714
- DOI: 10.3389/fcell.2022.899917
miRNA mediated downregulation of cyclase-associated protein 1 (CAP1) is required for myoblast fusion
Abstract
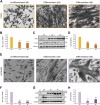
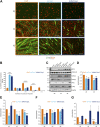
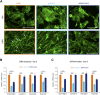
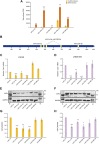
Myoblast fusion is essential for the formation, growth, and regeneration of skeletal muscle, but the molecular mechanisms that govern fusion and myofiber formation remain poorly understood. Past studies have shown an important role of the actin cytoskeleton and actin regulators in myoblast fusion. The Cyclase-Associated Proteins (CAP) 1 and 2 recently emerged as critical regulators of actin treadmilling in higher eukaryotes including mammals. Whilst the role of CAP2 in skeletal muscle development and function is well characterized, involvement of CAP1 in this process remains elusive. Here we report that CAP1, plays a critical role in cytoskeletal remodeling during myoblast fusion and formation of myotubes. Cap1 mRNA and protein are expressed in both murine C2C12 and human LHCN-M2 myoblasts, but their abundance decreases during myogenic differentiation. Perturbing the temporally controlled expression of CAP1 by overexpression or CRISPR-Cas9 mediated knockout impaired actin rearrangement, myoblast alignment, expression of profusion molecules, differentiation into multinucleated myotubes, and myosin heavy chain expression. Endogenous Cap1 expression is post-transcriptionally downregulated during differentiation by canonical myomiRs miR-1, miR-133, and miR-206, which have conserved binding sites at the 3' UTR of the Cap1 mRNA. Deletion of the endogenous 3' UTR by CRISPR-Cas9 in C2C12 cells phenocopies overexpression of CAP1 by inhibiting myotube formation. Our findings implicates Cap1 and its myomiR-mediated downregulation in the myoblast fusion process and the generation of skeletal muscle.
Keywords: CAP1; CAP2; microRNA; muscle differentiation; myogenesis; myosin heavy chain; post-transcriptional regulation.
Copyright © 2022 Singh, Rai, Weber and Posern.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Nap1-mediated actin remodeling is essential for mammalian myoblast fusion.J Cell Sci. 2009 Sep 15;122(Pt 18):3282-93. doi: 10.1242/jcs.047597. Epub 2009 Aug 25. J Cell Sci. 2009. PMID: 19706686 Free PMC article.
-
Stac3 inhibits myoblast differentiation into myotubes.PLoS One. 2014 Apr 30;9(4):e95926. doi: 10.1371/journal.pone.0095926. eCollection 2014. PLoS One. 2014. PMID: 24788338 Free PMC article.
-
Serum concentration impacts myosin heavy chain expression but not cellular respiration in human LHCN-M2 myoblasts undergoing differentiation.Exp Physiol. 2023 Feb;108(2):169-176. doi: 10.1113/EP090564. Epub 2023 Jan 9. Exp Physiol. 2023. PMID: 36621799 Free PMC article.
-
Mechanisms regulating myoblast fusion: A multilevel interplay.Semin Cell Dev Biol. 2020 Aug;104:81-92. doi: 10.1016/j.semcdb.2020.02.004. Epub 2020 Feb 13. Semin Cell Dev Biol. 2020. PMID: 32063453 Review.
-
Signaling mechanisms in mammalian myoblast fusion.Sci Signal. 2013 Apr 23;6(272):re2. doi: 10.1126/scisignal.2003832. Sci Signal. 2013. PMID: 23612709 Free PMC article. Review.
Cited by
-
Role of Actin-Binding Proteins in Skeletal Myogenesis.Cells. 2023 Oct 25;12(21):2523. doi: 10.3390/cells12212523. Cells. 2023. PMID: 37947600 Free PMC article. Review.
References
-
- Biederer C. H., Ries S. J., Moser M., Florio M., Israel M. A., McCormick F., et al. (2000). The basic helix-loop-helix transcription factors myogenin and Id2 mediate specific induction of caveolin-3 gene expression during embryonic development. J. Biol. Chem. 275 (34), 26245–26251. 10.1074/jbc.M001430200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

