DRESIS: the first comprehensive landscape of drug resistance information
- PMID: 36243960
- PMCID: PMC9825618
- DOI: 10.1093/nar/gkac812
DRESIS: the first comprehensive landscape of drug resistance information
Abstract
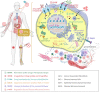
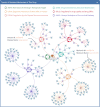
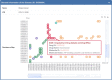
Widespread drug resistance has become the key issue in global healthcare. Extensive efforts have been made to reveal not only diverse diseases experiencing drug resistance, but also the six distinct types of molecular mechanisms underlying this resistance. A database that describes a comprehensive list of diseases with drug resistance (not just cancers/infections) and all types of resistance mechanisms is now urgently needed. However, no such database has been available to date. In this study, a comprehensive database describing drug resistance information named 'DRESIS' was therefore developed. It was introduced to (i) systematically provide, for the first time, all existing types of molecular mechanisms underlying drug resistance, (ii) extensively cover the widest range of diseases among all existing databases and (iii) explicitly describe the clinically/experimentally verified resistance data for the largest number of drugs. Since drug resistance has become an ever-increasing clinical issue, DRESIS is expected to have great implications for future new drug discovery and clinical treatment optimization. It is now publicly accessible without any login requirement at: https://idrblab.org/dresis/.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
TheMarker: a comprehensive database of therapeutic biomarkers.Nucleic Acids Res. 2024 Jan 5;52(D1):D1450-D1464. doi: 10.1093/nar/gkad862. Nucleic Acids Res. 2024. PMID: 37850638 Free PMC article.
-
NPCDR: natural product-based drug combination and its disease-specific molecular regulation.Nucleic Acids Res. 2022 Jan 7;50(D1):D1324-D1333. doi: 10.1093/nar/gkab913. Nucleic Acids Res. 2022. PMID: 34664659 Free PMC article.
-
DrugMAP: molecular atlas and pharma-information of all drugs.Nucleic Acids Res. 2023 Jan 6;51(D1):D1288-D1299. doi: 10.1093/nar/gkac813. Nucleic Acids Res. 2023. PMID: 36243961 Free PMC article.
-
Tuberculosis.In: Holmes KK, Bertozzi S, Bloom BR, Jha P, editors. Major Infectious Diseases. 3rd edition. Washington (DC): The International Bank for Reconstruction and Development / The World Bank; 2017 Nov 3. Chapter 11. In: Holmes KK, Bertozzi S, Bloom BR, Jha P, editors. Major Infectious Diseases. 3rd edition. Washington (DC): The International Bank for Reconstruction and Development / The World Bank; 2017 Nov 3. Chapter 11. PMID: 30212088 Free Books & Documents. Review.
-
Database systems for knowledge-based discovery.Methods Mol Biol. 2009;575:159-72. doi: 10.1007/978-1-60761-274-2_6. Methods Mol Biol. 2009. PMID: 19727614 Review.
Cited by
-
SingPro: a knowledge base providing single-cell proteomic data.Nucleic Acids Res. 2024 Jan 5;52(D1):D552-D561. doi: 10.1093/nar/gkad830. Nucleic Acids Res. 2024. PMID: 37819028 Free PMC article.
-
Dysregulated Signalling Pathways Driving Anticancer Drug Resistance.Int J Mol Sci. 2023 Jul 30;24(15):12222. doi: 10.3390/ijms241512222. Int J Mol Sci. 2023. PMID: 37569598 Free PMC article. Review.
-
TTD: Therapeutic Target Database describing target druggability information.Nucleic Acids Res. 2024 Jan 5;52(D1):D1465-D1477. doi: 10.1093/nar/gkad751. Nucleic Acids Res. 2024. PMID: 37713619 Free PMC article.
-
Identifying potential drug-target interactions based on ensemble deep learning.Front Aging Neurosci. 2023 Jun 15;15:1176400. doi: 10.3389/fnagi.2023.1176400. eCollection 2023. Front Aging Neurosci. 2023. PMID: 37396659 Free PMC article.
-
Multi-omics analysis reveals BZW1's regulation of EMT via the Wnt pathway in lung adenocarcinoma.J Cell Mol Med. 2024 Oct;28(20):e70163. doi: 10.1111/jcmm.70163. J Cell Mol Med. 2024. PMID: 39462268 Free PMC article.
References
-
- Kozic M., Fox S.J., Thomas J.M., Verma C.S., Rigden D.J.. Large scale ab initio modeling of structurally uncharacterized antimicrobial peptides reveals known and novel folds. Proteins. 2018; 86:548–565. - PubMed
-
- Bertagnolio S., Jordan M.R., Doherty M.. HIV drug resistance. N. Engl. J. Med. 2018; 378:874. - PubMed
-
- Mullard A. Stemming the tide of drug resistance in cancer. Nat. Rev. Drug Discov. 2020; 19:221–223. - PubMed
-
- Zhan J.Y., Ma J., Zheng Q.C.. Molecular dynamics investigation on the asciminib resistance mechanism of I502L and V468F mutations in BCR-ABL. J. Mol. Graph. Model. 2019; 89:242–249. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

