Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data
- PMID: 36235335
- PMCID: PMC9573011
- DOI: 10.3390/plants11192469
Identification of Key Genes Related to Dormancy Control in Prunus Species by Meta-Analysis of RNAseq Data
Abstract
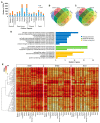
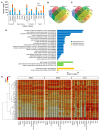
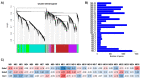
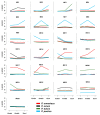
Bud dormancy is a genotype-dependent mechanism observed in Prunus species in which bud growth is inhibited, and the accumulation of a specific amount of chilling (endodormancy) and heat (ecodormancy) is necessary to resume growth and reach flowering. We analyzed publicly available transcriptome data from fifteen cultivars of four Prunus species (almond, apricot, peach, and sweet cherry) sampled at endo- and ecodormancy points to identify conserved genes and pathways associated with dormancy control in the genus. A total of 13,018 genes were differentially expressed during dormancy transitions, of which 139 and 223 were of interest because their expression profiles correlated with endo- and ecodormancy, respectively, in at least one cultivar of each species. The endodormancy-related genes comprised transcripts mainly overexpressed during chilling accumulation and were associated with abiotic stresses, cell wall modifications, and hormone regulation. The ecodormancy-related genes, upregulated after chilling fulfillment, were primarily involved in the genetic control of carbohydrate regulation, hormone biosynthesis, and pollen development. Additionally, the integrated co-expression network of differentially expressed genes in the four species showed clusters of co-expressed genes correlated to dormancy stages and genes of breeding interest overlapping with quantitative trait loci for bloom time and chilling and heat requirements.
Keywords: breeding; candidate genes; chilling and heat requirements; co-expression network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Distinctive Gene Expression Patterns Define Endodormancy to Ecodormancy Transition in Apricot and Peach.Front Plant Sci. 2020 Feb 28;11:180. doi: 10.3389/fpls.2020.00180. eCollection 2020. Front Plant Sci. 2020. PMID: 32180783 Free PMC article.
-
Meta-analysis of RNA-Seq studies reveals genes with dominant functions during flower bud endo- to eco-dormancy transition in Prunus species.Sci Rep. 2021 Jun 23;11(1):13173. doi: 10.1038/s41598-021-92600-6. Sci Rep. 2021. PMID: 34162991 Free PMC article.
-
Identification of early and late flowering time candidate genes in endodormant and ecodormant almond flower buds.Tree Physiol. 2021 Apr 8;41(4):589-605. doi: 10.1093/treephys/tpaa151. Tree Physiol. 2021. PMID: 33200186 Free PMC article.
-
Recent advancements to study flowering time in almond and other Prunus species.Front Plant Sci. 2014 Jul 11;5:334. doi: 10.3389/fpls.2014.00334. eCollection 2014. Front Plant Sci. 2014. PMID: 25071812 Free PMC article. Review.
-
Bud endodormancy in deciduous fruit trees: advances and prospects.Hortic Res. 2021 Jun 1;8(1):139. doi: 10.1038/s41438-021-00575-2. Hortic Res. 2021. PMID: 34078882 Free PMC article. Review.
References
-
- Lang G.A., Early J.D., Martin G.C., Darnell R.L. Endo-, para-and ecodormancy: Physiological terminology and classification for dormancy research. Hortic. Sci. 1987;22:271–277.
-
- Fadón E., Herrero M., Rodrigo J. Advances in Plant Dormancy. Springer; Berlin/Heidelberg, Germany: 2015. Flower bud dormancy in Prunus species; pp. 123–135. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

