Highly Conserved Interaction Profiles between Clinically Relevant Mutants of the Cytomegalovirus CDK-like Kinase pUL97 and Human Cyclins: Functional Significance of Cyclin H
- PMID: 36233116
- PMCID: PMC9569496
- DOI: 10.3390/ijms231911814
Highly Conserved Interaction Profiles between Clinically Relevant Mutants of the Cytomegalovirus CDK-like Kinase pUL97 and Human Cyclins: Functional Significance of Cyclin H
Abstract
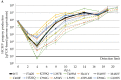
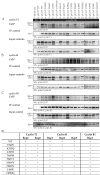
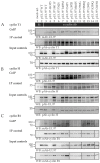
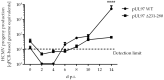
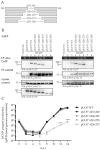
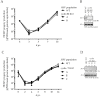
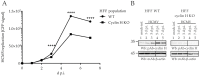
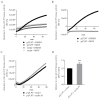
The complex host interaction network of human cytomegalovirus (HCMV) involves the regulatory protein kinase pUL97, which represents a viral cyclin-dependent kinase (CDK) ortholog. pUL97 interacts with the three human cyclin types T1, H, and B1, whereby the binding region of cyclin T1 and the pUL97 oligomerization region were both assigned to amino acids 231-280. We further addressed the question of whether HCMVs harboring mutations in ORF-UL97, i.e., short deletions or resistance-conferring point mutations, are affected in the interaction with human cyclins and viral replication. To this end, clinically relevant UL97 drug-resistance-conferring mutants were analyzed by whole-genome sequencing and used for genetic marker transfer experiments. The recombinant HCMVs indicated conservation of pUL97-cyclin interaction, since all viral UL97 point mutants continued to interact with the analyzed cyclin types and exerted wild-type-like replication fitness. In comparison, recombinant HCMVs UL97 Δ231-280 and also the smaller deletion Δ236-275, but not Δ241-270, lost interaction with cyclins T1 and H, showed impaired replication efficiency, and also exhibited reduced kinase activity. Moreover, a cellular knock-out of cyclins B1 or T1 did not alter HCMV replication phenotypes or pUL97 kinase activity, possibly indicating alternative, compensatory pUL97-cyclin interactions. In contrast, however, cyclin H knock-out, similar to virus deletion mutants in the pUL97-cyclin H binding region, exhibited strong defective phenotypes of HCMV replication, as supported by reduced pUL97 kinase activity in a cyclin H-dependent coexpression setting. Thus, cyclin H proved to be a very relevant determinant of pUL97 kinase activity and viral replication efficiency. As a conclusion, the results provide evidence for the functional importance of pUL97-cyclin interaction. High selective pressure on the formation of pUL97-cyclin complexes was identified by the use of clinically relevant mutants.
Keywords: clinically relevant viral mutants; cyclin H functional significance; functional relevance; human cyclin complexes; human cytomegalovirus; kinase activity; mapping and knock-out analyses; pUL97/vCDK; pUL97–cyclin interaction; viral CDK-like kinase; viral replication efficiency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Functional Relevance of the Interaction between Human Cyclins and the Cytomegalovirus-Encoded CDK-Like Protein Kinase pUL97.Viruses. 2021 Jun 27;13(7):1248. doi: 10.3390/v13071248. Viruses. 2021. PMID: 34198986 Free PMC article.
-
Cyclins B1, T1, and H differ in their molecular mode of interaction with cytomegalovirus protein kinase pUL97.J Biol Chem. 2019 Apr 12;294(15):6188-6203. doi: 10.1074/jbc.RA118.007049. Epub 2019 Feb 19. J Biol Chem. 2019. PMID: 30782840 Free PMC article.
-
Understanding the Cytomegalovirus Cyclin-Dependent Kinase Ortholog pUL97 as a Multifaceted Regulator and an Antiviral Drug Target.Cells. 2024 Aug 13;13(16):1338. doi: 10.3390/cells13161338. Cells. 2024. PMID: 39195228 Free PMC article. Review.
-
Cytomegalovirus cyclin-dependent kinase ortholog vCDK/pUL97 undergoes regulatory interaction with human cyclin H and CDK7 to codetermine viral replication efficiency.Virus Res. 2023 Oct 2;335:199200. doi: 10.1016/j.virusres.2023.199200. Epub 2023 Aug 19. Virus Res. 2023. PMID: 37591314 Free PMC article.
-
Regulatory roles of protein kinases in cytomegalovirus replication.Adv Virus Res. 2011;80:69-101. doi: 10.1016/B978-0-12-385987-7.00004-X. Adv Virus Res. 2011. PMID: 21762822 Review.
Cited by
-
An Antiherpesviral Host-Directed Strategy Based on CDK7 Covalently Binding Drugs: Target-Selective, Picomolar-Dose, Cross-Virus Reactivity.Pharmaceutics. 2024 Jan 23;16(2):158. doi: 10.3390/pharmaceutics16020158. Pharmaceutics. 2024. PMID: 38399219 Free PMC article.
-
Combined Treatment with Host-Directed and Anticytomegaloviral Kinase Inhibitors: Mechanisms, Synergisms and Drug Resistance Barriers.Pharmaceutics. 2023 Nov 27;15(12):2680. doi: 10.3390/pharmaceutics15122680. Pharmaceutics. 2023. PMID: 38140021 Free PMC article.
-
Assessment of Covalently Binding Warhead Compounds in the Validation of the Cytomegalovirus Nuclear Egress Complex as an Antiviral Target.Cells. 2023 Apr 14;12(8):1162. doi: 10.3390/cells12081162. Cells. 2023. PMID: 37190072 Free PMC article.
-
Cyclin-Dependent Kinase 8 Represents a Positive Regulator of Cytomegalovirus Replication and a Novel Host Target for Antiviral Strategies.Pharmaceutics. 2024 Sep 23;16(9):1238. doi: 10.3390/pharmaceutics16091238. Pharmaceutics. 2024. PMID: 39339274 Free PMC article.
-
The Interactive Complex between Cytomegalovirus Kinase vCDK/pUL97 and Host Factors CDK7-Cyclin H Determines Individual Patterns of Transcription in Infected Cells.Int J Mol Sci. 2023 Dec 13;24(24):17421. doi: 10.3390/ijms242417421. Int J Mol Sci. 2023. PMID: 38139252 Free PMC article.
References
-
- Goodrum F., Britt W., Mocarski E.S. Fields Virology: DNA Viruses. 7th ed. LWW; Philadelphia, PA, USA: 2021.
MeSH terms
Substances
Grants and funding
- IZKF project A88-M.M./H.S./Interdisciplinary Center of Clinical Research of the Medical Center/Universitätsklinikum Erlangen
- grant M.M./Ma.S./Matching Funds Program of the Forschungsstiftung Medizin, Medical Center UKER (Erlangen) together with the Manfred Roth-Stiftung (Fürth)
- grant DeeP CMV/AP-5/M.M./Bayerische Forschungsstiftung
- grants MM/WDR 2015-16, 2017-18, 2020-21/DAAD-Go8
LinkOut - more resources
Full Text Sources
Research Materials

