Subtractive genomic analysis for computational identification of putative immunogenic targets against clinical Enterobacter cloacae complex
- PMID: 36228013
- PMCID: PMC9560131
- DOI: 10.1371/journal.pone.0275749
Subtractive genomic analysis for computational identification of putative immunogenic targets against clinical Enterobacter cloacae complex
Abstract
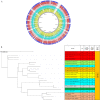
Background: Enterobacter is a major nosocomial genus of Enterobacteriaceae responsible for a variety of nosocomial infections, particularly in prolonged hospitalized patients in the intensive care units. Since current antibiotics have failed treating colistin- and carbapenem-resistant Enterobacteriaceae, efforts are underway to find suitable alternative strategies. Therefore, this study conducted a reverse vaccinology (RV) to identify novel and putative immunogenic targets using core proteome of 20 different sequence types (STs) of clinical Enterobacter spp. Moreover, we introduced a structural-based approach for exploration of potential vaccine candidates against the Enterobacteriaceae family using their conserved domain analysis.
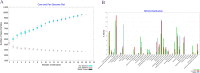
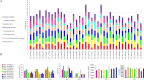
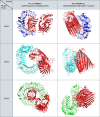
Results: A number of 2616 core coding sequences (CDSs) were retrieved from 20 clinical strains of Enterobacter spp. with a similarity of ≥ 50%. Nine proteins with a score of ≥ 20 considered as the shortlisted proteins based on the quartile scoring method, including three TonB-dependent receptors, WP_008500981.1, WP_058690971.1 and WP_058679571.1; one YjbH domain-containing protein, WP_110108068.1; three flagellar proteins, WP_088207510.1, WP_033145204.1 and WP_058679632.1; one spore-coat U domain-containing protein, WP_039266612.1; and one DD-metalloendopeptidase family protein, WP_025912449.1. In this study, proteins WP_058690971.1 and WP_110108068.1 were detected as the top candidates with regard to immune stimulation and interactions with TLRs. However, their efficacy is remaining to be evaluated experimentally.
Conclusions: Our investigation introduced common ferrichrome porins with high sequence similarity as potential vaccine candidates against the Enterobacteriaceae family. These proteins belong to the iron acquisition system and possess all criteria of suitable vaccine targets. Therefore, they need to be specifically paid attention for vaccine development against clinically important members of Enterobacteriaceae family.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Investigation of novel putative immunogenic targets against Staphylococcus aureus using a reverse vaccinology strategy.Infect Genet Evol. 2021 Dec;96:105149. doi: 10.1016/j.meegid.2021.105149. Epub 2021 Nov 18. Infect Genet Evol. 2021. PMID: 34801756
-
Identification of novel putative immunogenic targets and construction of a multi-epitope vaccine against multidrug-resistant Corynebacterium jeikeium using reverse vaccinology approach.Microb Pathog. 2022 Mar;164:105425. doi: 10.1016/j.micpath.2022.105425. Epub 2022 Jan 31. Microb Pathog. 2022. PMID: 35114352
-
Colistin Susceptibility in Companion Animal-Derived Escherichia coli, Klebsiella spp., and Enterobacter spp. in Japan: Frequent Isolation of Colistin-Resistant Enterobacter cloacae Complex.Front Cell Infect Microbiol. 2022 Jul 6;12:946841. doi: 10.3389/fcimb.2022.946841. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35873176 Free PMC article.
-
Vaccinomics to design a novel single chimeric subunit vaccine for broad-spectrum immunological applications targeting nosocomial Enterobacteriaceae pathogens.Eur J Pharm Sci. 2020 Apr 15;146:105258. doi: 10.1016/j.ejps.2020.105258. Epub 2020 Feb 5. Eur J Pharm Sci. 2020. PMID: 32035109
-
Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance.Future Microbiol. 2012 Jul;7(7):887-902. doi: 10.2217/fmb.12.61. Future Microbiol. 2012. PMID: 22827309 Review.
References
-
- Mishra M, Panda S, Barik S, Sarkar A, Singh DV, Mohapatra H. Antibiotic Resistance Profile, Outer Membrane Proteins, Virulence Factors and Genome Sequence Analysis Reveal Clinical Isolates of Enterobacter Are Potential Pathogens Compared to Environmental Isolates. Front Cell Infect Microbiol. 2020;10:54. Epub 20200221. doi: 10.3389/fcimb.2020.00054 ; PubMed Central PMCID: PMC7047878. - DOI - PMC - PubMed
-
- Ji Y, Wang P, Xu T, Zhou Y, Chen R, Zhu H, et al.. Development of a One-Step Multiplex PCR Assay for Differential Detection of Four species (Enterobacter cloacae, Enterobacter hormaechei, Enterobacter roggenkampii, and Enterobacter kobei) Belonging to Enterobacter cloacae Complex With Clinical Significance. Front Cell Infect Microbiol. 2021;11:677089. Epub 20210518. doi: 10.3389/fcimb.2021.677089 ; PubMed Central PMCID: PMC8169972. - DOI - PMC - PubMed
-
- Xu T, Zhang C, Ji Y, Song J, Liu Y, Guo Y, et al.. Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ Pollut. 2021;268(Pt B):115706. Epub 20201012. doi: 10.1016/j.envpol.2020.115706 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

