Allelic variation of Escherichia coli outer membrane protein A: Impact on cell surface properties, stress tolerance and allele distribution
- PMID: 36227900
- PMCID: PMC9560509
- DOI: 10.1371/journal.pone.0276046
Allelic variation of Escherichia coli outer membrane protein A: Impact on cell surface properties, stress tolerance and allele distribution
Abstract
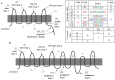
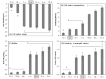
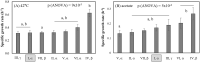
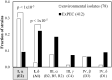
Outer membrane protein A (OmpA) is one of the most abundant outer membrane proteins of Gram-negative bacteria and is known to have patterns of sequence variations at certain amino acids-allelic variation-in Escherichia coli. Here we subjected seven exemplar OmpA alleles expressed in a K-12 (MG1655) ΔompA background to further characterization. These alleles were observed to significantly impact cell surface charge (zeta potential), cell surface hydrophobicity, biofilm formation, sensitivity to killing by neutrophil elastase, and specific growth rate at 42°C and in the presence of acetate, demonstrating that OmpA is an attractive target for engineering cell surface properties and industrial phenotypes. It was also observed that cell surface charge and biofilm formation both significantly correlate with cell surface hydrophobicity, a cell property that is increasingly intriguing for bioproduction. While there was poor alignment between the observed experimental values relative to the known sequence variation, differences in hydrophobicity and biofilm formation did correspond to the identity of residue 203 (N vs T), located within the proposed dimerization domain. The relative abundance of the (I, δ) allele was increased in extraintestinal pathogenic E. coli (ExPEC) isolates relative to environmental isolates, with a corresponding decrease in (I, α) alleles in ExPEC relative to environmental isolates. The (I, α) and (I, δ) alleles differ at positions 203 and 251. Variations in distribution were also observed among ExPEC types and phylotypes. Thus, OmpA allelic variation and its influence on OmpA function warrant further investigation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
OmpA of a septicemic Escherichia coli O78--secretion and convergent evolution.Int J Med Microbiol. 2004 Oct;294(6):373-81. doi: 10.1016/j.ijmm.2004.08.004. Int J Med Microbiol. 2004. PMID: 15595387
-
Outer membrane protein A (OmpA) of extraintestinal pathogenic Escherichia coli.BMC Res Notes. 2020 Jan 31;13(1):51. doi: 10.1186/s13104-020-4917-5. BMC Res Notes. 2020. PMID: 32005127 Free PMC article.
-
Peptides Affecting the Outer Membrane Lipid Asymmetry System (MlaA-OmpC/F) Reduce Avian Pathogenic Escherichia coli (APEC) Colonization in Chickens.Appl Environ Microbiol. 2021 Aug 11;87(17):e0056721. doi: 10.1128/AEM.00567-21. Epub 2021 Aug 11. Appl Environ Microbiol. 2021. PMID: 34132592 Free PMC article.
-
Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase.Science. 2000 Aug 18;289(5482):1185-8. doi: 10.1126/science.289.5482.1185. Science. 2000. PMID: 10947984
-
Secreted proteases: A new insight in the pathogenesis of extraintestinal pathogenic Escherichia coli.Int J Med Microbiol. 2019 May-Jun;309(3-4):159-168. doi: 10.1016/j.ijmm.2019.03.002. Epub 2019 Mar 22. Int J Med Microbiol. 2019. PMID: 30940425 Review.
Cited by
-
Outer Membrane Porins Contribute to Antimicrobial Resistance in Gram-Negative Bacteria.Microorganisms. 2023 Jun 28;11(7):1690. doi: 10.3390/microorganisms11071690. Microorganisms. 2023. PMID: 37512863 Free PMC article. Review.
References
-
- Jarboe LR, Klauda JB, Chen Y, Davis KM, Santoscoy MC. Engineering the Microbial Cell Membrane To Improve Bioproduction. In: Cheng HN, Gross RA, Smith PB, editors. Green Polymer Chemistry: New Products, Processes, and Applications. ACS Symposium Series. . p. 25–39.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

