The origins of COVID-19 pandemic: A brief overview
- PMID: 36218169
- PMCID: PMC9874793
- DOI: 10.1111/tbed.14732
The origins of COVID-19 pandemic: A brief overview
Abstract
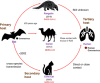
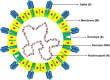
The novel coronavirus disease (COVID-19) outbreak that emerged at the end of 2019 has now swept the world for more than 2 years, causing immeasurable damage to the lives and economies of the world. It has drawn so much attention to discovering how the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) originated and entered the human body. The current argument revolves around two contradictory theories: a scenario of laboratory spillover events and human contact with zoonotic diseases. Here, we reviewed the transmission, pathogenesis, possible hosts, as well as the genome and protein structure of SARS-CoV-2, which play key roles in the COVID-19 pandemic. We believe the coronavirus was originally transmitted to human by animals rather than by a laboratory leak. However, there still needs more investigations to determine the source of the pandemic. Understanding how COVID-19 emerged is vital to developing global strategies for mitigating future outbreaks.
Keywords: SARS-CoV-2; genome similarities; laboratory origin; protein structure evolution; zoonotic origin.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare that the work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

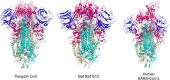
Similar articles
-
The novel zoonotic COVID-19 pandemic: An expected global health concern.J Infect Dev Ctries. 2020 Mar 31;14(3):254-264. doi: 10.3855/jidc.12671. J Infect Dev Ctries. 2020. PMID: 32235085
-
Zoonotic origins of human coronaviruses.Int J Biol Sci. 2020 Mar 15;16(10):1686-1697. doi: 10.7150/ijbs.45472. eCollection 2020. Int J Biol Sci. 2020. PMID: 32226286 Free PMC article. Review.
-
Human and novel coronavirus infections in children: a review.Paediatr Int Child Health. 2021 Feb;41(1):36-55. doi: 10.1080/20469047.2020.1781356. Epub 2020 Jun 25. Paediatr Int Child Health. 2021. PMID: 32584199 Review.
-
SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights.J Basic Microbiol. 2021 Mar;61(3):180-202. doi: 10.1002/jobm.202000537. Epub 2021 Jan 18. J Basic Microbiol. 2021. PMID: 33460172 Free PMC article. Review.
-
Viral, host and environmental factors that favor anthropozoonotic spillover of coronaviruses: An opinionated review, focusing on SARS-CoV, MERS-CoV and SARS-CoV-2.Sci Total Environ. 2021 Jan 1;750:141483. doi: 10.1016/j.scitotenv.2020.141483. Epub 2020 Aug 5. Sci Total Environ. 2021. PMID: 32829257 Free PMC article. Review.
Cited by
-
Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.iScience. 2024 Jan 19;27(2):108976. doi: 10.1016/j.isci.2024.108976. eCollection 2024 Feb 16. iScience. 2024. PMID: 38327783 Free PMC article.
-
Chloroquine: Rapidly withdrawing from first-line treatment of COVID-19.Heliyon. 2024 Aug 28;10(17):e37098. doi: 10.1016/j.heliyon.2024.e37098. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39281655 Free PMC article. Review.
-
Advances in the Search for SARS-CoV-2 Mpro and PLpro Inhibitors.Pathogens. 2024 Sep 24;13(10):825. doi: 10.3390/pathogens13100825. Pathogens. 2024. PMID: 39452697 Free PMC article. Review.
-
Editorial: Wildlife-domestic animal interface: threat or sentinel?Front Vet Sci. 2024 Sep 24;11:1495580. doi: 10.3389/fvets.2024.1495580. eCollection 2024. Front Vet Sci. 2024. PMID: 39380777 Free PMC article. No abstract available.
-
Lung Involvement Patterns in COVID-19: CT Scan Insights and Prognostic Implications From a Tertiary Care Center in Southern India.Cureus. 2024 Jan 31;16(1):e53335. doi: 10.7759/cureus.53335. eCollection 2024 Jan. Cureus. 2024. PMID: 38435896 Free PMC article.
References
-
- Amanat, F. , Thapa, M. , Lei, T. , Ahmed, S. M. S. , Adelsberg, D. C. , Carreno, J. M. , Strohmeier, S. , Schmitz, A. J. , Zafar, S. , Zhou, J. Q. , Rijnink, W. , Alshammary, H. , Borcherding, N. , Reiche, A. G. , Srivastava, K. , Sordillo, E. M. , van Bakel, H. , Personalized Virology, I. , Turner, J. S. , & Krammer, F. (2021). SARS‐CoV‐2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell, 184(15), 3936–3948. e3910. 10.1016/j.cell.2021.06.005 - DOI - PMC - PubMed
-
- Amendola, A. , Bianchi, S. , Gori, M. , Colzani, D. , Canuti, M. , Borghi, E. , Raviglione, M. C. , Zuccotti, G. V. , & Tanzi, E. (2021). Evidence of SARS‐CoV‐2 RNA in an oropharyngeal swab specimen, Milan, Italy, early December 2019. Emerging Infectious Diseases, 27(2), 648–650. 10.3201/eid2702.204632 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 2020XM01/Key Medical Science and Technology Program of Shanxi Province
- XD1809/Doctor of Shanxi Medical University
- SD1809/Science Research Start-up Fund for Doctor of Shanxi Province
- 2019L0414/Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi
- 201901D111189/Applied Basic Research Program of Shanxi Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

