Substitutions near the HA receptor binding site explain the origin and major antigenic change of the B/Victoria and B/Yamagata lineages
- PMID: 36215486
- PMCID: PMC9586307
- DOI: 10.1073/pnas.2211616119
Substitutions near the HA receptor binding site explain the origin and major antigenic change of the B/Victoria and B/Yamagata lineages
Abstract
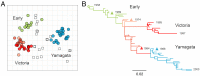
Influenza B virus primarily infects humans, causing seasonal epidemics globally. Two antigenic variants-Victoria-like and Yamagata-like-were detected in the 1980s, of which the molecular basis of emergence is still incompletely understood. Here, the antigenic properties of a unique collection of historical virus isolates, sampled from 1962 to 2000 and passaged exclusively in mammalian cells to preserve antigenic properties, were determined with the hemagglutination inhibition assay and an antigenic map was built to quantify and visualize the divergence of the lineages. The antigenic map revealed only three distinct antigenic clusters-Early, Victoria, and Yamagata-with relatively little antigenic diversity in each cluster until 2000. Viruses with Victoria-like antigenic properties emerged around 1972 and diversified subsequently into two genetic lineages. Viruses with Yamagata-like antigenic properties evolved from one lineage and became clearly antigenically distinct from the Victoria-like viruses around 1988. Recombinant mutant viruses were tested to show that insertions and deletions (indels), as observed frequently in influenza B virus hemagglutinin, had little effect on antigenic properties. In contrast, amino-acid substitutions at positions 148, 149, 150, and 203, adjacent to the hemagglutinin receptor binding site, determined the main antigenic differences between the Early, Victoria-like, and Yamagata-like viruses. Surprisingly, substitutions at two of the four positions reverted in recent viruses of the Victoria lineage, resulting in antigenic properties similar to viruses circulating ∼50 y earlier. These data shed light on the antigenic diversification of influenza viruses and suggest there may be limits to the antigenic evolution of influenza B virus.
Keywords: antigenic evolution; hemagglutinin; influenza B virus; vaccines.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Molecular characterization and evolution dynamics of influenza B viruses circulating in Germany from season 1996/1997 to 2019/2020.Virus Res. 2022 Dec;322:198926. doi: 10.1016/j.virusres.2022.198926. Epub 2022 Sep 10. Virus Res. 2022. PMID: 36096395
-
[The Victoria and Yamagata Lineages of Influenza B Viruses, unknown and undervalued].Rev Esp Quimioter. 2022 Jun;35(3):231-235. doi: 10.37201/req/159.2021. Epub 2022 Feb 18. Rev Esp Quimioter. 2022. PMID: 35180825 Free PMC article. Review. Spanish.
-
Changes in the hemagglutinins and neuraminidases of human influenza B viruses isolated in Italy during the 2001-02, 2002-03, and 2003-04 seasons.J Med Virol. 2004 Dec;74(4):629-40. doi: 10.1002/jmv.20225. J Med Virol. 2004. PMID: 15484280
-
Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):619-628. doi: 10.1073/pnas.1916585116. Epub 2019 Dec 16. Proc Natl Acad Sci U S A. 2020. PMID: 31843889 Free PMC article.
-
Recent changes among human influenza viruses.Virus Res. 2004 Jul;103(1-2):47-52. doi: 10.1016/j.virusres.2004.02.011. Virus Res. 2004. PMID: 15163487 Review.
Cited by
-
Co-evolution of immunity and seasonal influenza viruses.Nat Rev Microbiol. 2023 Dec;21(12):805-817. doi: 10.1038/s41579-023-00945-8. Epub 2023 Aug 2. Nat Rev Microbiol. 2023. PMID: 37532870 Review.
-
Age-dependent influenza infection patterns and subtype circulation in Denmark, in seasons 2015/16 to 2021/22.Euro Surveill. 2024 Jan;29(4):2300263. doi: 10.2807/1560-7917.ES.2024.29.4.2300263. Euro Surveill. 2024. PMID: 38275020 Free PMC article.
-
Whole-Genome Analysis of Influenza A(H3N2) and B/Victoria Viruses Detected in Myanmar during the COVID-19 Pandemic in 2021.Viruses. 2023 Feb 20;15(2):583. doi: 10.3390/v15020583. Viruses. 2023. PMID: 36851797 Free PMC article.
-
Immune imprinting in early life shapes cross-reactivity to influenza B virus haemagglutinin.Nat Microbiol. 2024 Aug;9(8):2073-2083. doi: 10.1038/s41564-024-01732-8. Epub 2024 Jun 18. Nat Microbiol. 2024. PMID: 38890491
-
Triton X-100-treated virus-based ELLA demonstrates discordant antigenic evolution of influenza B virus hemagglutinin and neuraminidase.J Virol. 2024 Oct 22;98(10):e0118624. doi: 10.1128/jvi.01186-24. Epub 2024 Oct 3. J Virol. 2024. PMID: 39360825
References
-
- T. Francis, Jr, A new type of virus from epidemic influenza. Science 92, 405–408 (1940). - PubMed
-
- Rota P. A., et al. , Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175, 59–68 (1990). - PubMed
-
- Chen J.-M., et al. , Exploration of the emergence of the Victoria lineage of influenza B virus. Arch. Virol. 152, 415–422 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

