Development of a versatile high-throughput mutagenesis assay with multiplexed short-read NGS using DNA-barcoded supF shuttle vector library amplified in E. coli
- PMID: 36214452
- PMCID: PMC9584611
- DOI: 10.7554/eLife.83780
Development of a versatile high-throughput mutagenesis assay with multiplexed short-read NGS using DNA-barcoded supF shuttle vector library amplified in E. coli
Abstract
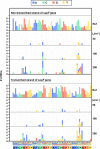
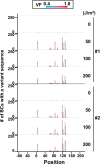
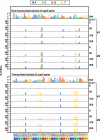
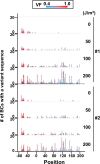
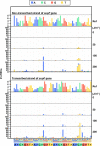
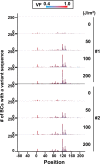
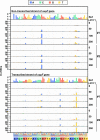
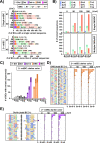
A forward mutagenesis assay using the supF gene has been widely employed for the last several decades in studies addressing mutation frequencies and mutation spectra associated with various intrinsic and environmental mutagens. In this study, by using a supF shuttle vector and non-SOS-induced Escherichia coli with short-read next-generation sequencing (NGS) technology, we present an advanced method for the study of mutations, which is simple, versatile, and cost-effective. We demonstrate the performance of our newly developed assay via pilot experiments with ultraviolet (UV) irradiation, the results from which emerge more relevant than expected. The NGS data obtained from samples of the indicator E. coli grown on titer plates provides mutation frequency and spectrum data, and uncovers obscure mutations that cannot be detected by a conventional supF assay. Furthermore, a very small amount of NGS data from selection plates reveals the almost full spectrum of mutations in each specimen and offers us a novel insight into the mechanisms of mutagenesis, despite them being considered already well known. We believe that the method presented here will contribute to future opportunities for research on mutagenesis, DNA repair, and cancer.
Keywords: NGS; UV mutagenesis; cell biology; mutagenesis assay; none; supF.
© 2022, Kawai et al.
Conflict of interest statement
HK, RI, SE, RS, SM, CF, SK, HK No competing interests declared
Figures
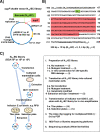
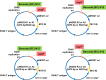

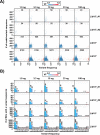


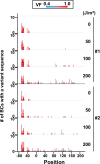






Similar articles
-
A method for selection of forward mutations in supF gene carried by shuttle-vector plasmids.Carcinogenesis. 1993 Feb;14(2):303-5. doi: 10.1093/carcin/14.2.303. Carcinogenesis. 1993. PMID: 8435871
-
An Escherichia coli plasmid-based, mutational system in which supF mutants are selectable: insertion elements dominate the spontaneous spectra.Mutat Res. 1992 Nov 16;270(2):219-31. doi: 10.1016/0027-5107(92)90133-m. Mutat Res. 1992. PMID: 1383739
-
Mutational specificity of 1-(2-chloroethyl)-3-cyclohexyl-1-nitrosourea in Escherichia coli: comparison of in vivo with in vitro exposure of the supF gene.Environ Mol Mutagen. 1997;30(1):65-71. doi: 10.1002/(sici)1098-2280(1997)30:1<65::aid-em9>3.0.co;2-b. Environ Mol Mutagen. 1997. PMID: 9258331
-
Mutation spectra in supF: approaches to elucidating sequence context effects.Mutat Res. 2000 May 30;450(1-2):61-73. doi: 10.1016/s0027-5107(00)00016-6. Mutat Res. 2000. PMID: 10838134 Review.
-
Mutational spectra for polycyclic aromatic hydrocarbons in the supF target gene.Mutat Res. 2000 May 30;450(1-2):75-93. doi: 10.1016/s0027-5107(00)00017-8. Mutat Res. 2000. PMID: 10838135 Review.
Cited by
-
Flow-cytometric cell sorting coupled with UV mutagenesis for improving pectin lyase expression.Front Bioeng Biotechnol. 2023 Aug 31;11:1251342. doi: 10.3389/fbioe.2023.1251342. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37720319 Free PMC article.
References
-
- Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, PCAWG Mutational Signatures Working Group. Getz G, Rozen SG, Stratton MR, PCAWG Consortium The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

