Cancer-associated fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in lung adenocarcinoma
- PMID: 36209111
- PMCID: PMC9548164
- DOI: 10.1186/s13045-022-01359-4
Cancer-associated fibroblast-specific lncRNA LINC01614 enhances glutamine uptake in lung adenocarcinoma
Abstract
Background: Besides featured glucose consumption, recent studies reveal that cancer cells might prefer "addicting" specific energy substrates from the tumor microenvironment (TME); however, the underlying mechanisms remain unclear.
Methods: Fibroblast-specific long noncoding RNAs were screened using RNA-seq data of our NJLCC cohort, TCGA, and CCLE datasets. The expression and package of LINC01614 into exosomes were identified using flow cytometric sorting, fluorescence in situ hybridization (FISH), and quantitative reverse transcription polymerase chain reaction (RT-PCR). The transfer and functional role of LINC01614 in lung adenocarcinoma (LUAD) and CAFs were investigated using 4-thiouracil-labeled RNA transfer and gain- and loss-of-function approaches. RNA pull-down, RNA immunoprecipitation, dual-luciferase assay, gene expression microarray, and bioinformatics analysis were performed to investigate the underlying mechanisms involved.
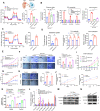
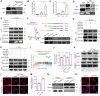
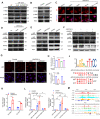
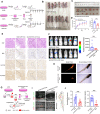
Results: We demonstrate that cancer-associated fibroblasts (CAFs) in LUAD primarily enhance the glutamine metabolism of cancer cells. A CAF-specific long noncoding RNA, LINC01614, packaged by CAF-derived exosomes, mediates the enhancement of glutamine uptake in LUAD cells. Mechanistically, LINC01614 directly interacts with ANXA2 and p65 to facilitate the activation of NF-κB, which leads to the upregulation of the glutamine transporters SLC38A2 and SLC7A5 and eventually enhances the glutamine influx of cancer cells. Reciprocally, tumor-derived proinflammatory cytokines upregulate LINC01614 in CAFs, constituting a feedforward loop between CAFs and cancer cells. Blocking exosome-transmitted LINC01614 inhibits glutamine addiction and LUAD growth in vivo. Clinically, LINC01614 expression in CAFs is associated with the glutamine influx and poor prognosis of patients with LUAD.
Conclusion: Our study highlights the therapeutic potential of targeting a CAF-specific lncRNA to inhibit glutamine utilization and cancer progression in LUAD.
Keywords: Cancer-associated fibroblasts; Glutamine; Long noncoding RNA; Metabolic reprograming; Tumor microenvironment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Identification of a novel therapeutic candidate, NRK, in primary cancer-associated fibroblasts of lung adenocarcinoma microenvironment.J Cancer Res Clin Oncol. 2021 Apr;147(4):1049-1064. doi: 10.1007/s00432-020-03489-z. Epub 2021 Jan 2. J Cancer Res Clin Oncol. 2021. PMID: 33387038
-
Cancer-associated fibroblasts-derived exosomal METTL3 promotes the proliferation, invasion, stemness and glutaminolysis in non-small cell lung cancer cells by eliciting SLC7A5 m6A modification.Hum Cell. 2024 Jul;37(4):1120-1131. doi: 10.1007/s13577-024-01056-z. Epub 2024 Apr 16. Hum Cell. 2024. PMID: 38625505
-
Knockdown of LINC01614 inhibits lung adenocarcinoma cell progression by up-regulating miR-217 and down-regulating FOXP1.J Cell Mol Med. 2018 Sep;22(9):4034-4044. doi: 10.1111/jcmm.13483. Epub 2018 Jun 22. J Cell Mol Med. 2018. PMID: 29934982 Free PMC article.
-
Dissecting the roles of exosomal cancer-associated fibroblasts-derived non-coding RNAs in tumor progression: A complete guide.Pathol Res Pract. 2024 Oct;262:155576. doi: 10.1016/j.prp.2024.155576. Epub 2024 Aug 30. Pathol Res Pract. 2024. PMID: 39232286 Review.
-
The promising role of noncoding RNAs in cancer-associated fibroblasts: an overview of current status and future perspectives.J Hematol Oncol. 2020 Nov 19;13(1):154. doi: 10.1186/s13045-020-00988-x. J Hematol Oncol. 2020. PMID: 33213510 Free PMC article. Review.
Cited by
-
Cancer associated fibroblasts and metabolic reprogramming: unraveling the intricate crosstalk in tumor evolution.J Hematol Oncol. 2024 Sep 2;17(1):80. doi: 10.1186/s13045-024-01600-2. J Hematol Oncol. 2024. PMID: 39223656 Free PMC article. Review.
-
The biological function of tumor-derived extracellular vesicles on metabolism.Cell Commun Signal. 2023 Jun 22;21(1):150. doi: 10.1186/s12964-023-01111-6. Cell Commun Signal. 2023. PMID: 37349803 Free PMC article. Review.
-
A risk model based on ferroptosis- and cuproptosis-related lncRNAs predicts prognosis and immune microenvironment in lung adenocarcinoma by bioinformatics analysis and experimental verification.Am J Cancer Res. 2023 Nov 15;13(11):5306-5319. eCollection 2023. Am J Cancer Res. 2023. PMID: 38058804 Free PMC article.
-
Integrating single-cell and bulk RNA sequencing to develop a cancer-associated fibroblast-related signature for immune infiltration prediction and prognosis in lung adenocarcinoma.J Thorac Dis. 2023 Mar 31;15(3):1406-1425. doi: 10.21037/jtd-23-238. J Thorac Dis. 2023. PMID: 37065583 Free PMC article.
-
[Latest Findings on Long Noncoding RNA in Tumor Microenvironment].Sichuan Da Xue Xue Bao Yi Xue Ban. 2023 May;54(3):491-496. doi: 10.12182/20230560507. Sichuan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37248573 Free PMC article. Review. Chinese.
References
-
- Dey P, Kimmelman AC, DePinho RA. Metabolic codependencies in the tumor microenvironment. Cancer Discov. 2021;11(5):1067–1081. doi: 10.1158/2159-8290.CD-20-1211. - DOI - PMC - PubMed
-
- Wang M. Preferential glutamine uptake in cancer cells. Nat Rev Nephrol. 2021;17(6):368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

