The RNA m6A writer WTAP in diseases: structure, roles, and mechanisms
- PMID: 36207306
- PMCID: PMC9546849
- DOI: 10.1038/s41419-022-05268-9
The RNA m6A writer WTAP in diseases: structure, roles, and mechanisms
Abstract
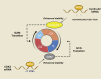
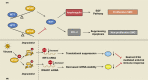
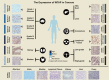
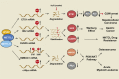
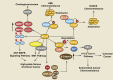
N6-methyladenosine (m6A) is a widely investigated RNA modification in studies on the "epigenetic regulation" of mRNAs that is ubiquitously present in eukaryotes. Abnormal changes in m6A levels are closely related to the regulation of RNA metabolism, heat shock stress, tumor occurrence, and development. m6A modifications are catalyzed by the m6A writer complex, which contains RNA methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), Wilms tumor 1-associated protein (WTAP), and other proteins with methyltransferase (MTase) capability, such as RNA-binding motif protein 15 (RBM15), KIAA1429 and zinc finger CCCH-type containing 13 (ZC3H13). Although METTL3 is the main catalytic subunit, WTAP is a regulatory subunit whose function is to recruit the m6A methyltransferase complex to the target mRNA. Specifically, WTAP is required for the accumulation of METTL3 and METTL14 in nuclear speckles. In this paper, we briefly introduce the molecular mechanism of m6A modification. Then, we focus on WTAP, a component of the m6A methyltransferase complex, and introduce its structure, localization, and physiological functions. Finally, we describe its roles and mechanisms in cancer.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The RNA m6A writer METTL14 in cancers: Roles, structures, and applications.Biochim Biophys Acta Rev Cancer. 2021 Dec;1876(2):188609. doi: 10.1016/j.bbcan.2021.188609. Epub 2021 Aug 8. Biochim Biophys Acta Rev Cancer. 2021. PMID: 34375716 Review.
-
Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase.Cell Res. 2014 Feb;24(2):177-89. doi: 10.1038/cr.2014.3. Epub 2014 Jan 10. Cell Res. 2014. PMID: 24407421 Free PMC article.
-
WTAP Targets the METTL3 m6A-Methyltransferase Complex to Cytoplasmic Hepatitis C Virus RNA to Regulate Infection.J Virol. 2022 Nov 23;96(22):e0099722. doi: 10.1128/jvi.00997-22. Epub 2022 Oct 31. J Virol. 2022. PMID: 36314819 Free PMC article.
-
METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP.Cell Biol Int. 2020 Dec;44(12):2524-2531. doi: 10.1002/cbin.11459. Epub 2020 Sep 11. Cell Biol Int. 2020. PMID: 32869897
-
Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential.Biomolecules. 2022 Sep 2;12(9):1224. doi: 10.3390/biom12091224. Biomolecules. 2022. PMID: 36139062 Free PMC article. Review.
Cited by
-
Unraveling the landscape of m6A RNA methylation in wound healing and scars.Cell Death Discov. 2024 Oct 29;10(1):458. doi: 10.1038/s41420-024-02222-w. Cell Death Discov. 2024. PMID: 39472463 Free PMC article. Review.
-
Autophagy: a critical mechanism of N6-methyladenosine modification involved in tumor progression and therapy resistance.Cell Death Dis. 2024 Oct 28;15(10):783. doi: 10.1038/s41419-024-07148-w. Cell Death Dis. 2024. PMID: 39468015 Free PMC article. Review.
-
The role of KRT7 in metastasis and prognosis of pancreatic cancer.Cancer Cell Int. 2024 Sep 19;24(1):321. doi: 10.1186/s12935-024-03500-4. Cancer Cell Int. 2024. PMID: 39300449 Free PMC article.
-
The m6A regulators in prostate cancer: molecular basis and clinical perspective.Front Pharmacol. 2024 Aug 29;15:1448872. doi: 10.3389/fphar.2024.1448872. eCollection 2024. Front Pharmacol. 2024. PMID: 39268470 Free PMC article. Review.
-
Icariin ameliorates TNF-α/IFN-γ-induced oxidative stress, inflammatory response and apoptosis of human immortalized epidermal cells through the WTAP/SERPINB4 axis.Arch Dermatol Res. 2024 Aug 23;316(8):557. doi: 10.1007/s00403-024-03281-w. Arch Dermatol Res. 2024. PMID: 39177922
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

