Phagocytosing differentiated cell-fragments is a novel mechanism for controlling somatic stem cell differentiation within a short time frame
- PMID: 36203068
- PMCID: PMC9537123
- DOI: 10.1007/s00018-022-04555-0
Phagocytosing differentiated cell-fragments is a novel mechanism for controlling somatic stem cell differentiation within a short time frame
Abstract
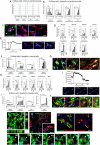
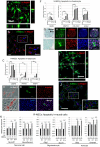
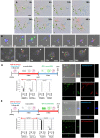
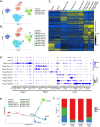
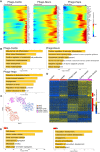
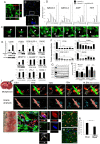
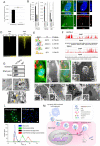
Stem cells undergo cytokine-driven differentiation, but this process often takes longer than several weeks to complete. A novel mechanism for somatic stem cell differentiation via phagocytosing 'model cells' (apoptotic differentiated cells) was found to require only a short time frame. Pluripotent-like Muse cells, multipotent mesenchymal stem cells (MSCs), and neural stem cells (NSCs) phagocytosed apoptotic differentiated cells via different phagocytic receptor subsets than macrophages. The phagocytosed-differentiated cell-derived contents (e.g., transcription factors) were quickly released into the cytoplasm, translocated into the nucleus, and bound to promoter regions of the stem cell genomes. Within 24 ~ 36 h, the cells expressed lineage-specific markers corresponding to the phagocytosed-differentiated cells, both in vitro and in vivo. At 1 week, the gene expression profiles were similar to those of the authentic differentiated cells and expressed functional markers. Differentiation was limited to the inherent potential of each cell line: triploblastic-, adipogenic-/chondrogenic-, and neural-lineages in Muse cells, MSCs, and NSCs, respectively. Disruption of phagocytosis, either by phagocytic receptor inhibition via small interfering RNA or annexin V treatment, impeded differentiation in vitro and in vivo. Together, our findings uncovered a simple mechanism by which differentiation-directing factors are directly transferred to somatic stem cells by phagocytosing apoptotic differentiated cells to trigger their rapid differentiation into the target cell lineage.
Keywords: Apoptotic cells; Differentiation; Muse cells; Phagosomal release; Single-cell RNA sequencing; Somatic stem cells.
© 2022. The Author(s).
Conflict of interest statement
S. Wakao, Y. Kushida, Y. Kuroda, and M. Dezawa are affiliated with Tohoku University Graduate School of Medicine and are parties to a co-development agreement with Life Science Institute, Inc. (LSII; Tokyo, Japan). S. Wakao and M. Dezawa have a patent for Muse cells, and the isolation method thereof is licensed to LSII.
Figures
Similar articles
-
Naïve pluripotent-like characteristics of non-tumorigenic Muse cells isolated from human amniotic membrane.Sci Rep. 2022 Oct 14;12(1):17222. doi: 10.1038/s41598-022-22282-1. Sci Rep. 2022. PMID: 36241699 Free PMC article.
-
A matter of identity - Phenotype and differentiation potential of human somatic stem cells.Stem Cell Res. 2015 Jul;15(1):1-13. doi: 10.1016/j.scr.2015.04.003. Epub 2015 Apr 25. Stem Cell Res. 2015. PMID: 25957945
-
Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes.PLoS One. 2012;7(7):e40399. doi: 10.1371/journal.pone.0040399. Epub 2012 Jul 12. PLoS One. 2012. PMID: 22808153 Free PMC article.
-
Mesenchymal stem cells and their subpopulation, pluripotent muse cells, in basic research and regenerative medicine.Anat Rec (Hoboken). 2014 Jan;297(1):98-110. doi: 10.1002/ar.22798. Epub 2013 Dec 2. Anat Rec (Hoboken). 2014. PMID: 24293378 Review.
-
Muse Cells Provide the Pluripotency of Mesenchymal Stem Cells: Direct Contribution of Muse Cells to Tissue Regeneration.Cell Transplant. 2016;25(5):849-61. doi: 10.3727/096368916X690881. Epub 2016 Feb 15. Cell Transplant. 2016. PMID: 26884346 Review.
Cited by
-
Human post-implantation blastocyst-like characteristics of Muse cells isolated from human umbilical cord.Cell Mol Life Sci. 2024 Jul 11;81(1):297. doi: 10.1007/s00018-024-05339-4. Cell Mol Life Sci. 2024. PMID: 38992309 Free PMC article.
-
Intravenous Administration of Human Muse Cells Ameliorates Deficits in a Rat Model of Subacute Spinal Cord Injury.Int J Mol Sci. 2023 Sep 27;24(19):14603. doi: 10.3390/ijms241914603. Int J Mol Sci. 2023. PMID: 37834052 Free PMC article.
-
Human Muse cells isolated from preterm- and term-umbilical cord delivered therapeutic effects in rat bleomycin-induced lung injury model without immunosuppressant.Stem Cell Res Ther. 2024 May 22;15(1):147. doi: 10.1186/s13287-024-03763-8. Stem Cell Res Ther. 2024. PMID: 38773627 Free PMC article.
-
Investigating the Potential of Multilineage Differentiating Stress-Enduring Cells for Osteochondral Healing.Cartilage. 2024 Jun 17:19476035241262020. doi: 10.1177/19476035241262020. Online ahead of print. Cartilage. 2024. PMID: 38887038 Free PMC article.
-
Stem Cell Therapy for Acute/Subacute Ischemic Stroke with a Focus on Intraarterial Stem Cell Transplantation: From Basic Research to Clinical Trials.Bioengineering (Basel). 2022 Dec 27;10(1):33. doi: 10.3390/bioengineering10010033. Bioengineering (Basel). 2022. PMID: 36671605 Free PMC article. Review.
References
-
- Gimeno ML, Fuertes F, BarcalaTabarrozzi AE, Attorressi AI, Cucchiani R, Corrales L, Oliveira TC, Sogayar MC, Labriola L, Dewey RA, Perone MJ. Pluripotent nontumorigenic adipose tissue-derived muse cells have immunomodulatory capacity mediated by transforming growth factor-β1. Stem Cells Transl Med. 2017;6:161–173. doi: 10.5966/sctm.2016-0014. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

