Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance
- PMID: 36200462
- PMCID: PMC10942761
- DOI: 10.1039/d2cc04541a
Beyond darunavir: recent development of next generation HIV-1 protease inhibitors to combat drug resistance
Abstract
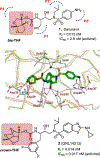
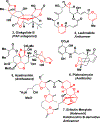
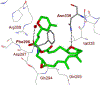
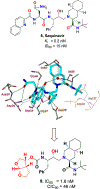
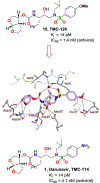
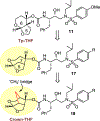
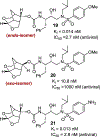
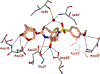
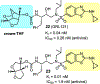
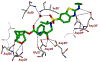
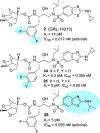
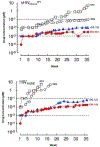
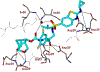
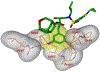
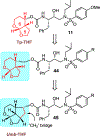
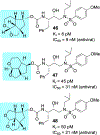
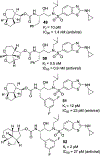
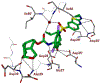
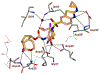
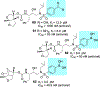
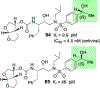
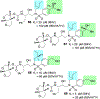
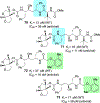
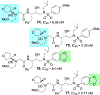
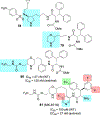
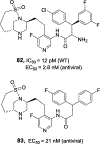
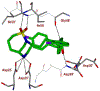
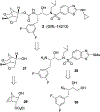
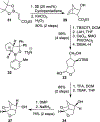
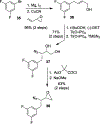
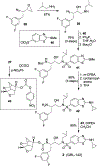
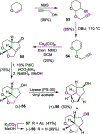
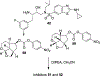
We report our recent development of a conceptually new generation of exceptionally potent non-peptidic HIV-1 protease inhibitors that displayed excellent pharmacological and drug-resistance profiles. Our X-ray structural studies of darunavir and other designed inhibitors from our laboratories led us to create a variety of inhibitors incorporating fused ring polycyclic ethers and aromatic heterocycles to promote hydrogen bonding interactions with the backbone atoms of HIV-1 protease as well as van der Waals interactions with residues in the S2 and S2' subsites. We have also incorporated specific functionalities to enhance van der Waals interactions in the S1 and S1' subsites. The combined effects of these structural templates are critical to the inhibitors' exceptional potency and drug-like properties. We highlight here our molecular design strategies to promote backbone hydrogen bonding interactions to combat drug-resistance and specific design of polycyclic ether templates to mimic peptide-like bonds in the HIV-1 protease active site. Our medicinal chemistry and drug development efforts led to the development of new generation inhibitors significantly improved over darunavir and displaying unprecedented antiviral activity against multidrug-resistant HIV-1 variants.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures
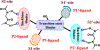

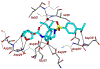
Similar articles
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Nature Inspired Molecular Design: Stereoselective Synthesis of Bicyclic and Polycyclic Ethers for Potent HIV-1 Protease Inhibitors.Asian J Org Chem. 2018 Aug;7(8):1448-1466. doi: 10.1002/ajoc.201800255. Epub 2018 Jun 8. Asian J Org Chem. 2018. PMID: 31595212 Free PMC article.
-
Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.ChemMedChem. 2017 Dec 7;12(23):1942-1952. doi: 10.1002/cmdc.201700614. Epub 2017 Nov 24. ChemMedChem. 2017. PMID: 29110408 Free PMC article.
-
Design, synthesis, protein-ligand X-ray structure, and biological evaluation of a series of novel macrocyclic human immunodeficiency virus-1 protease inhibitors to combat drug resistance.J Med Chem. 2009 Dec 10;52(23):7689-705. doi: 10.1021/jm900695w. J Med Chem. 2009. PMID: 19746963 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.Bioorg Med Chem Lett. 2023 Mar 1;83:129168. doi: 10.1016/j.bmcl.2023.129168. Epub 2023 Feb 3. Bioorg Med Chem Lett. 2023. PMID: 36738797 Free PMC article.
-
HIV-1 protease inhibitors with a P1 phosphonate modification maintain potency against drug-resistant variants by increased interactions with flap residues.Eur J Med Chem. 2023 Sep 5;257:115501. doi: 10.1016/j.ejmech.2023.115501. Epub 2023 May 18. Eur J Med Chem. 2023. PMID: 37244161 Free PMC article.
-
Recent Advances on Targeting Proteases for Antiviral Development.Viruses. 2024 Feb 27;16(3):366. doi: 10.3390/v16030366. Viruses. 2024. PMID: 38543732 Free PMC article. Review.
-
SARS-CoV-2 Mpro inhibitor identification using a cellular gain-of-signal assay for high-throughput screening.SLAS Discov. 2024 Sep;29(6):100181. doi: 10.1016/j.slasd.2024.100181. Epub 2024 Aug 22. SLAS Discov. 2024. PMID: 39173830 Free PMC article.
-
Extracellular Vesicle-Based SARS-CoV-2 Vaccine.Vaccines (Basel). 2023 Feb 24;11(3):539. doi: 10.3390/vaccines11030539. Vaccines (Basel). 2023. PMID: 36992123 Free PMC article. Review.
References
-
- Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong MAWP, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, Tiemessen CT, Vandekerckhove L and Lewin SR, The International AIDS Society (IAS) Global Scientific Strategy working group, Nature Med. 2021, 27, 2085–2098. - PubMed
-
- Frieden TR, Foti KE and Mermin J, N. Engl. J. Med 2015, 373, 2281–2287. - PubMed
-
- UNAIDS/WHO. Global HIV/AIDS Response - Progress Report 2021, November 2021. Available at http://www.unaids.org/en/resources/publications/2011/, Last accessed May 2012.
-
- de Clercq E, Int. J. Antimicrob. Agents 2009, 33, 307–320. - PubMed
-
- Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ and Holmberg SD, New Engl. J. Med 1998, 338, 853–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

