Super-Resolution Microscopy of the Synaptonemal Complex within the Caenorhabditis elegans Germline
- PMID: 36190293
- PMCID: PMC7614930
- DOI: 10.3791/64363
Super-Resolution Microscopy of the Synaptonemal Complex within the Caenorhabditis elegans Germline
Abstract
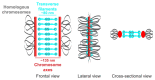
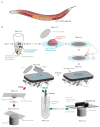
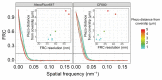
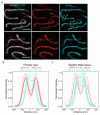
During meiosis, homologous chromosomes must recognize and adhere to one another to allow for their correct segregation. One of the key events that secures the interaction of homologous chromosomes is the assembly of the synaptonemal complex (SC) in meiotic prophase I. Even though there is little sequence homology between protein components within the SC among different species, the general structure of the SC has been highly conserved during evolution. In electron micrographs, the SC appears as a tripartite, ladder-like structure composed of lateral elements or axes, transverse filaments, and a central element. However, precisely identifying the localization of individual components within the complex by electron microscopy to determine the molecular structure of the SC remains challenging. By contrast, fluorescence microscopy allows for the identification of individual protein components within the complex. However, since the SC is only ~100 nm wide, its substructure cannot be resolved by diffraction-limited conventional fluorescence microscopy. Thus, determining the molecular architecture of the SC requires super-resolution light microscopy techniques such as structured illumination microscopy (SIM), stimulated-emission depletion (STED) microscopy, or single-molecule localization microscopy (SMLM). To maintain the structure and interactions of individual components within the SC, it is important to observe the complex in an environment that is close to its native environment in the germ cells. Therefore, we demonstrate an immunohistochemistry and imaging protocol that enables the study of the substructure of the SC in intact, extruded Caenorhabditis elegans germline tissue with SMLM and STED microscopy. Directly fixing the tissue to the coverslip reduces the movement of the samples during imaging and minimizes aberrations in the sample to achieve the high resolution necessary to visualize the substructure of the SC in its biological context.
Conflict of interest statement
Authors declare no conflicts of interest.
Figures


Similar articles
-
CRL4 regulates recombination and synaptonemal complex aggregation in the Caenorhabditis elegans germline.PLoS Genet. 2019 Nov 18;15(11):e1008486. doi: 10.1371/journal.pgen.1008486. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31738749 Free PMC article.
-
Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue.Proc Natl Acad Sci U S A. 2017 Jun 13;114(24):E4734-E4743. doi: 10.1073/pnas.1702312114. Epub 2017 May 30. Proc Natl Acad Sci U S A. 2017. PMID: 28559338 Free PMC article.
-
CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis.PLoS Genet. 2008 Jun 6;4(6):e1000088. doi: 10.1371/journal.pgen.1000088. PLoS Genet. 2008. PMID: 18535664 Free PMC article.
-
The Mammalian synaptonemal complex: a scaffold and beyond.Genome Dyn. 2009;5:69-80. doi: 10.1159/000166620. Genome Dyn. 2009. PMID: 18948708 Review.
-
The synaptonemal complex of basal metazoan hydra: more similarities to vertebrate than invertebrate meiosis model organisms.J Genet Genomics. 2014 Mar 20;41(3):107-15. doi: 10.1016/j.jgg.2014.01.009. Epub 2014 Feb 20. J Genet Genomics. 2014. PMID: 24656231 Review.
Cited by
-
Skp1 proteins are structural components of the synaptonemal complex in C. elegans.Sci Adv. 2024 Feb 16;10(7):eadl4876. doi: 10.1126/sciadv.adl4876. Epub 2024 Feb 14. Sci Adv. 2024. PMID: 38354250 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
