Neuroblasts contribute to oligodendrocytes generation upon demyelination in the adult mouse brain
- PMID: 36185360
- PMCID: PMC9519617
- DOI: 10.1016/j.isci.2022.105102
Neuroblasts contribute to oligodendrocytes generation upon demyelination in the adult mouse brain
Abstract
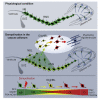
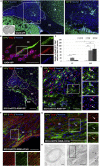
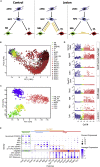
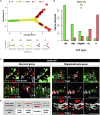
After demyelinating insult, the neuronal progenitors of the adult mouse sub-ventricular zone (SVZ) called neuroblasts convert into oligodendrocytes that participate to the remyelination process. We use this rare example of spontaneous fate conversion to identify the molecular mechanisms governing these processes. Using in vivo cell lineage and single cell RNA-sequencing, we demonstrate that SVZ neuroblasts fate conversion proceeds through formation of a non-proliferating transient cellular state co-expressing markers of both neuronal and oligodendrocyte identities. Transition between the two identities starts immediately after demyelination and occurs gradually, by a stepwise upregulation/downregulation of key TFs and chromatin modifiers. Each step of this fate conversion involves fine adjustments of the transcription and translation machineries as well as tight regulation of metabolism and migratory behaviors. Together, these data constitute the first in-depth analysis of a spontaneous cell fate conversion in the adult mammalian CNS.
Keywords: Cellular neuroscience; Developmental neuroscience; Molecular neuroscience.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Transient hypothyroidism favors oligodendrocyte generation providing functional remyelination in the adult mouse brain.Elife. 2017 Sep 6;6:e29996. doi: 10.7554/eLife.29996. Elife. 2017. PMID: 28875931 Free PMC article.
-
Promoting Myelin Repair through In Vivo Neuroblast Reprogramming.Stem Cell Reports. 2018 May 8;10(5):1492-1504. doi: 10.1016/j.stemcr.2018.02.015. Epub 2018 Mar 29. Stem Cell Reports. 2018. PMID: 29606615 Free PMC article.
-
Neural Stem Cells of the Subventricular Zone Contribute to Neuroprotection of the Corpus Callosum after Cuprizone-Induced Demyelination.J Neurosci. 2019 Jul 10;39(28):5481-5492. doi: 10.1523/JNEUROSCI.0227-18.2019. Epub 2019 May 28. J Neurosci. 2019. PMID: 31138656 Free PMC article.
-
The role of SVZ-derived neural precursors in demyelinating diseases: from animal models to multiple sclerosis.J Neurol Sci. 2008 Feb 15;265(1-2):26-31. doi: 10.1016/j.jns.2007.09.032. Epub 2007 Oct 24. J Neurol Sci. 2008. PMID: 17961598 Review.
-
The role of oligodendrocytes and oligodendrocyte progenitors in CNS remyelination.Adv Exp Med Biol. 1999;468:183-97. doi: 10.1007/978-1-4615-4685-6_15. Adv Exp Med Biol. 1999. PMID: 10635029 Review.
References
LinkOut - more resources
Full Text Sources

