P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7
- PMID: 36176277
- PMCID: PMC9513069
- DOI: 10.3389/fcell.2022.993525
P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7
Erratum in
-
Corrigendum: P14AS upregulates gene expression in the CDKN2A/2B locus through competitive binding to PcG protein CBX7.Front Cell Dev Biol. 2022 Dec 9;10:1062931. doi: 10.3389/fcell.2022.1062931. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36568974 Free PMC article.
Abstract
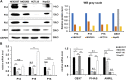
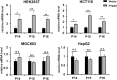
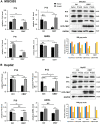
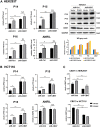
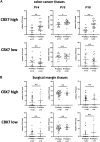
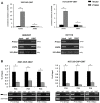
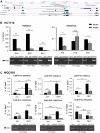
Background: It is well known that P16 INK4A , P14 ARF , P15 INK4B mRNAs, and ANRIL lncRNA are transcribed from the CDKN2A/2B locus. LncRNA P14AS is a lncRNA transcribed from antisense strand of P14 ARF promoter to intron-1. Our previous study showed that P14AS could upregulate the expression level of ANRIL and P16 INK4A and promote the proliferation of cancer cells. Because polycomb group protein CBX7 could repress P16 INK4A expression and bind ANRIL, we wonder whether the P14AS-upregulated ANRIL and P16 INK4A expression is mediated with CBX7. Results: In this study, we found that the upregulation of P16 INK4A , P14 ARF , P15 INK4B and ANRIL expression was induced by P14AS overexpression only in HEK293T and HCT116 cells with active endogenous CBX7 expression, but not in MGC803 and HepG2 cells with weak CBX7 expression. Further studies showed that the stable shRNA-knockdown of CBX7 expression abolished the P14AS-induced upregulation of these P14AS target genes in HEK293T and HCT116 cells whereas enforced CBX7 overexpression enabled P14AS to upregulate expression of these target genes in MGC803 and HepG2 cells. Moreover, a significant association between the expression levels of P14AS and its target genes were observed only in human colon cancer tissue samples with high level of CBX7 expression (n = 38, p < 0.05), but not in samples (n = 37) with low level of CBX7 expression, nor in paired surgical margin tissues. In addition, the results of RNA immunoprecipitation (RIP)- and chromatin immunoprecipitation (ChIP)-PCR analyses revealed that lncRNA P14AS could competitively bind to CBX7 protein which prevented the bindings of CBX7 to both lncRNA ANRIL and the promoters of P16 INK4A , P14 ARF and P15 INK4B genes. The amounts of repressive histone modification H3K9m3 was also significantly decreased at the promoters of these genes by P14AS in CBX7 actively expressing cells. Conclusions: CBX7 expression is essential for P14AS to upregulate the expression of P16 INK4A , P14 ARF , P15 INK4B and ANRIL genes in the CDKN2A/2Blocus. P14AS may upregulate these genes' expression through competitively blocking CBX7-binding to ANRIL lncRNA and target gene promoters.
Keywords: CBX7; P14AS; lncRNA; p14ARF; p15INK4B; p16INK4A.
Copyright © 2022 Li, Qiao, Ma, Zhou, Gu, Deng and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Characterization of novel LncRNA P14AS as a protector of ANRIL through AUF1 binding in human cells.Mol Cancer. 2020 Feb 27;19(1):42. doi: 10.1186/s12943-020-01150-4. Mol Cancer. 2020. PMID: 32106863 Free PMC article.
-
Expression of ANRIL-Polycomb Complexes-CDKN2A/B/ARF Genes in Breast Tumors: Identification of a Two-Gene (EZH2/CBX7) Signature with Independent Prognostic Value.Mol Cancer Res. 2016 Jul;14(7):623-33. doi: 10.1158/1541-7786.MCR-15-0418. Epub 2016 Apr 21. Mol Cancer Res. 2016. PMID: 27102007
-
Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression.Cancer Res. 2011 Aug 15;71(16):5365-9. doi: 10.1158/0008-5472.CAN-10-4379. Epub 2011 Aug 9. Cancer Res. 2011. PMID: 21828241 Free PMC article. Review.
-
Expression of Chr9p21 genes CDKN2B (p15(INK4b)), CDKN2A (p16(INK4a), p14(ARF)) and MTAP in human atherosclerotic plaque.Atherosclerosis. 2011 Feb;214(2):264-70. doi: 10.1016/j.atherosclerosis.2010.06.029. Epub 2010 Jun 23. Atherosclerosis. 2011. PMID: 20637465
-
Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health.Epigenetics. 2010 Nov-Dec;5(8):685-90. doi: 10.4161/epi.5.8.12996. Epub 2010 Nov 1. Epigenetics. 2010. PMID: 20716961 Free PMC article. Review.
Cited by
-
The linear ANRIL transcript P14AS regulates the NF-κB signaling to promote colon cancer progression.Mol Med. 2023 Dec 1;29(1):162. doi: 10.1186/s10020-023-00761-z. Mol Med. 2023. PMID: 38041015 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

