Microtubule integrity regulates budding yeast RAM pathway gene expression
- PMID: 36172269
- PMCID: PMC9511886
- DOI: 10.3389/fcell.2022.989820
Microtubule integrity regulates budding yeast RAM pathway gene expression
Abstract
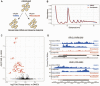
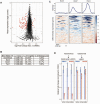
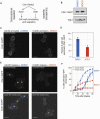
During mitosis, cells must spatiotemporally regulate gene expression programs to ensure accurate cellular division. Failures to properly regulate mitotic progression result in aneuploidy, a hallmark of cancer. Entry and exit from mitosis is largely controlled by waves of cyclin-dependent kinase (CDK) activity coupled to targeted protein degradation. The correct timing of CDK-based mitotic regulation is coordinated with the structure and function of microtubules. To determine whether mitotic gene expression is also regulated by the integrity of microtubules, we performed ribosome profiling and mRNA-sequencing in the presence and absence of microtubules in the budding yeast Saccharomyces cerevisiae. We discovered a coordinated translational and transcriptional repression of genes involved in cell wall biology processes when microtubules are disrupted. The genes targeted for repression in the absence of microtubules are enriched for downstream targets of a feed-forward pathway that controls cytokinesis and septum degradation and is regulated by the Cbk1 kinase, the Regulation of Ace2 Morphogenesis (RAM) pathway. We demonstrate that microtubule disruption leads to aberrant subcellular localization of Cbk1 in a manner that partially depends on the spindle position checkpoint. Furthermore, constitutive activation of the RAM pathway in the absence of microtubules leads to growth defects. Taken together, these results uncover a previously unknown link between microtubule function and the proper execution of mitotic gene expression programs to ensure that cell division does not occur prematurely.
Keywords: Cbk1; RAM pathway; microtubule; mitosis; ribosome profiling; transcription; translation.
Copyright © 2022 Lee and Biggins.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis.Mol Cell Biol. 2011 Feb;31(4):721-35. doi: 10.1128/MCB.00403-10. Epub 2010 Dec 6. Mol Cell Biol. 2011. PMID: 21135117 Free PMC article.
-
The Ndr/LATS Kinase Cbk1 Regulates a Specific Subset of Ace2 Functions and Suppresses the Hypha-to-Yeast Transition in Candida albicans.mBio. 2020 Aug 18;11(4):e01900-20. doi: 10.1128/mBio.01900-20. mBio. 2020. PMID: 32817109 Free PMC article.
-
The spindle position checkpoint requires positional feedback from cytoplasmic microtubules.Curr Biol. 2009 Dec 15;19(23):2026-30. doi: 10.1016/j.cub.2009.10.020. Epub 2009 Nov 12. Curr Biol. 2009. PMID: 19913426 Free PMC article.
-
MEN, destruction and separation: mechanistic links between mitotic exit and cytokinesis in budding yeast.Bioessays. 2002 Jul;24(7):659-66. doi: 10.1002/bies.10106. Bioessays. 2002. PMID: 12111726 Review.
-
Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1.Small GTPases. 2015 Oct 2;6(4):196-201. doi: 10.1080/21541248.2015.1109023. Small GTPases. 2015. PMID: 26507466 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

